Caveolin-1 Impacts on TGF-β Regulation of Metabolic Gene Signatures in Hepatocytes
- PMID: 32082178
- PMCID: PMC7005071
- DOI: 10.3389/fphys.2019.01606
Caveolin-1 Impacts on TGF-β Regulation of Metabolic Gene Signatures in Hepatocytes
Abstract
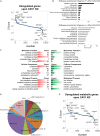
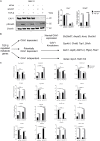
Caveolin-1 (CAV1) is a membrane protein associated with metabolism in various cell types. The transforming growth factor beta (TGF-β) is a pro-fibrogenic cytokine in the liver, but its metabolic gene signatures remain unclear to date. We have previously shown that CAV1 alters TGF-β signaling and blocks its pro-apoptotic function. Here, we defined TGF-β-induced metabolic gene signatures in hepatocytes and assessed whether CAV1 abundance affects TGF-β control of those metabolic genes. Microarray analyses of primary hepatocytes after TGF-β stimulation (48 h) showed differential expression of 4224 genes, of which 721 are metabolic genes (adjusted p < 0.001). Functional annotation analysis revealed that TGF-β mainly suppresses metabolic gene network, including genes involved in glutathione, cholesterol, fatty acid, and amino acid metabolism. TGF-β also upregulated several genes related to glycan metabolism and ion transport. In contrast to TGF-β effects, CAV1 knockdown triggered the upregulation of metabolic genes. Immortalized mouse hepatocytes (AML12 cells) were used to validate the gene changes induced by TGF-β stimulation and CAV1 knockdown. Noteworthy, of the TGF-β metabolic target genes, CAV1 modulated the expression of 228 (27%). In conclusion, we present several novel metabolic gene signatures of TGF-β in hepatocytes and show that CAV1 abundance alters almost a third of these genes. These findings could enable a better understanding of TGF-β function in normal and diseased liver especially where differential CAV1 level is implicated.
Keywords: caveolin-1; liver diseases; metabolism; microarray; transforming growth factor beta.
Copyright © 2020 Han, Nwosu, Piorońska, Ebert, Dooley and Meyer.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

