Cross-Continental Dispersal of Major HIV-1 CRF01_AE Clusters in China
- PMID: 32082287
- PMCID: PMC7005055
- DOI: 10.3389/fmicb.2020.00061
Cross-Continental Dispersal of Major HIV-1 CRF01_AE Clusters in China
Abstract
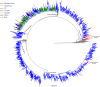
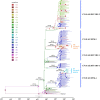
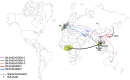
Since the 1990s, several distinct clusters of human immunodeficiency virus-type 1 (HIV-1) CRF01_AE related to a large epidemic in China have been identified, but it is yet poorly understood whether its transmission has dispersed globally. We aimed to characterize and quantify the genetic relationship of HIV-1 CRF01_AEs circulating in China and other countries. Using representative sequences of Chinese clusters as queries, all relevant CRF01_AE pol sequences in two large databases (the Los Alamos HIV sequence database and the UK HIV Drug Resistance Database) were selected with the online basic local alignment search (BLAST) tool. Phylogenetic and phylogeographic analyses were then carried out to characterize possible linkage of CRF01_AE strains between China and the rest of the world. We identified that 269 strains isolated in other parts of the world were associated with five major Chinese CRF01_AE clusters. 80.7% were located within CN.01AE.HST/IDU-2, most of which were born in Southeast Asia. 17.8% were clustered with CN.01AE.MSM-4 and -5. Two distinct sub-clusters associated with Chinese men who have sex with men (MSM) emerged in HK-United Kingdom and Japan after 2000. Our analysis suggests that HIV-1 CRF01_AE strains related to viral transmission in China were initially brought to the United Kingdom or other countries during the 1990s by Asian immigrants or returning international tourists from Southeast Asia, and then after having circulated among MSM in China for several years, these Chinese strains dispersed outside again, possibly through MSM network. This study provided evidence of regional and global dispersal of Chinese CRF01_AE strains. It would also help understand the global landscape of HIV epidemic associated with CRF01_AE transmission and highlight the need for further international collaborative study in this field.
Keywords: CRF01_AE; China; HIV-1; cross-continental; dispersal.
Copyright © 2020 An, Han, Zhao, English, Frost, Zhang and Shang.
Figures
References
-
- Angelis K., Albert J., Mamais I., Magiorkinis G., Hatzakis A., Hamouda O. (2015). Global dispersal pattern of HIV Type 1 Subtype CRF01_AE: a genetic trace of human mobility related to heterosexual sexual activities centralized in Southeast Asia. J. Infect. Dis. 211 1735–1744. 10.1093/infdis/jiu666 - DOI - PubMed
-
- Cuevas M., Fernandez-Garcia A., Sanchez-Garcia A., Gonzalez-Galeano M., Pinilla M., Sanchez-Martinez M. (2009). Incidence of non-B subtypes of HIV-1 in Galicia, Spain: high frequency and diversity of HIV-1 among men who have sex with men. Euro. Surveill. 14:19413. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

