COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis
- PMID: 32083328
- PMCID: PMC7228367
- DOI: 10.1002/jmv.25719
COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis
Abstract
Last December 2019, a new virus, named novel Coronavirus (COVID-2019) causing many cases of severe pneumonia was reported in Wuhan, China. The virus knowledge is limited and especially about COVID-2019 pathogenesis. The Open Reading Frame 1ab (ORF1ab) of COVID-2019 has been analyzed to evidence the presence of mutation caused by selective pressure on the virus. For selective pressure analysis fast-unconstrained Bayesian approximation (FUBAR) was used. Homology modelling has been performed by SwissModel and HHPred servers. The presence of transmembrane helical segments in Coronavirus ORF1ab non structural protein 2 (nsp2) and nsp3 was tested by TMHMM, MEMSAT, and MEMPACK tools. Three-dimensional structures have been analyzed and displayed using PyMOL. FUBAR analysis revealed the presence of potential sites under positive selective pressure (P < .05). Position 723 in the COVID-2019 has a serine instead a glycine residue, while at aminoacidic position 1010 a proline instead an isoleucine. Significant (P < .05) pervasive negative selection in 2416 sites (55%) was found. The positive selective pressure could account for some clinical features of this virus compared with severe acute respiratory syndrome (SARS) and Bat SARS-like CoV. The stabilizing mutation falling in the endosome-associated-protein-like domain of the nsp2 protein could account for COVID-2019 high ability of contagious, while the destabilizing mutation in nsp3 proteins could suggest a potential mechanism differentiating COVID-2019 from SARS. These data could be helpful for further investigation aimed to identify potential therapeutic targets or vaccine strategy, especially in the actual moment when the epidemic is ongoing and the scientific community is trying to enrich knowledge about this new viral pathogen.
Keywords: epidemiology; infection; pandemics; pathogenesis; protein-protein interaction analysis; research and analysis methods.
© 2020 Wiley Periodicals, Inc.
Figures
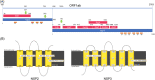
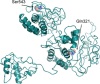
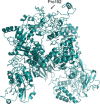
Comment in
-
Role of nonstructural proteins in the pathogenesis of SARS-CoV-2.J Med Virol. 2020 Sep;92(9):1427-1429. doi: 10.1002/jmv.25858. Epub 2020 Jun 2. J Med Virol. 2020. PMID: 32270884 Free PMC article. No abstract available.
References
-
- Hall TA. BioEdit A user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95‐98.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

