Deletion of Endonuclease V suppresses chemically induced hepatocellular carcinoma
- PMID: 32083667
- PMCID: PMC7192598
- DOI: 10.1093/nar/gkaa115
Deletion of Endonuclease V suppresses chemically induced hepatocellular carcinoma
Abstract
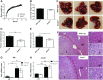
Endonuclease V (EndoV) is a conserved inosine-specific ribonuclease with unknown biological function. Here, we present the first mouse model lacking EndoV, which is viable without visible abnormalities. We show that endogenous murine EndoV cleaves inosine-containing RNA in vitro, nevertheless a series of experiments fails to link an in vivo function to processing of such transcripts. As inosine levels and adenosine-to-inosine editing often are dysregulated in hepatocellular carcinoma (HCC), we chemically induced HCC in mice. All mice developed liver cancer, however, EndoV-/- tumors were significantly fewer and smaller than wild type tumors. Opposed to human HCC, adenosine deaminase mRNA expression and site-specific editing were unaltered in our model. Loss of EndoV did not affect editing levels in liver tumors, however mRNA expression of a selection of cancer related genes were reduced. Inosines are also found in certain tRNAs and tRNAs are cleaved during stress to produce signaling entities. tRNA fragmentation was dysregulated in EndoV-/- livers and apparently, inosine-independent. We speculate that the inosine-ribonuclease activity of EndoV is disabled in vivo, but RNA binding allowed to promote stabilization of transcripts or recruitment of proteins to fine-tune gene expression. The EndoV-/- tumor suppressive phenotype calls for related studies in human HCC.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
-
- Gray M.W. Evolutionary origin of RNA editing. Biochemistry. 2012; 51:5235–5242. - PubMed
-
- Alseth I., Dalhus B., Bjoras M.. Inosine in DNA and RNA. Curr. Opin. Genet. Dev. 2014; 26:116–123. - PubMed
-
- Maas S., Rich A.. Changing genetic information through RNA editing. BioEssays: News Rev. Mol. Cell Dev. Biol. 2000; 22:790–802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

