Autophagic Degradation of NBR1 Restricts Metastatic Outgrowth during Mammary Tumor Progression
- PMID: 32084360
- PMCID: PMC7108946
- DOI: 10.1016/j.devcel.2020.01.025
Autophagic Degradation of NBR1 Restricts Metastatic Outgrowth during Mammary Tumor Progression
Abstract
Although autophagy is being pursued as a therapeutic target in clinical oncology trials, its effects on metastasis, the principal cause of cancer mortality, remain unclear. Here, we utilize mammary cancer models to temporally delete essential autophagy regulators during carcinoma progression. Though genetic ablation of autophagy strongly attenuates primary mammary tumor growth, impaired autophagy promotes spontaneous metastasis and enables the outgrowth of disseminated tumor cells into overt macro-metastases. Transcriptomic analysis reveals that autophagy deficiency elicits a subpopulation of otherwise luminal tumor cells exhibiting basal differentiation traits, which is reversed upon preventing accumulation of the autophagy cargo receptor, Neighbor to BRCA1 (NBR1). Furthermore, pharmacological and genetic induction of autophagy suppresses pro-metastatic differentiation and metastatic outgrowth. Analysis of human breast cancer data reveal that autophagy gene expression inversely correlates with pro-metastatic differentiation signatures and predicts overall and distant metastasis-free survival. Overall, these findings highlight autophagy-dependent control of NBR1 as a key determinant of metastatic progression.
Keywords: Keratin14; NBR1; Rubicon; TP63; autophagy; breast cancer; chloroquine; metastasis.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.D. is a scientific advisory board member for Vescor Therapeutics, LLC. Z.W. is a scientific advisory board member for Maverick Therapeutics, Inc.
Figures
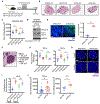

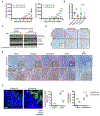



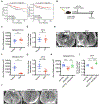
Comment in
-
Autophagy Suppresses Breast Cancer Metastasis.Dev Cell. 2020 Mar 9;52(5):542-544. doi: 10.1016/j.devcel.2020.02.005. Dev Cell. 2020. PMID: 32155434
References
-
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNAseq quantification. Nat Biotechnol 34, 525–527. - PubMed
-
- Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, Chambers AF, and MacDonald IC (2000). Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res 60, 2541–2546. - PubMed
-
- Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta KJ, Bader JS, et al. (2016). Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A 113, E854–863. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

