Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells
- PMID: 32084390
- PMCID: PMC7147146
- DOI: 10.1016/j.stem.2019.12.011
Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells
Abstract
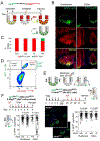
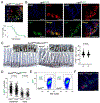
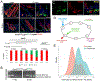
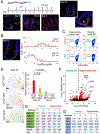
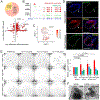
Ablation of LGR5+ intestinal stem cells (ISCs) is associated with rapid restoration of the ISC compartment. Different intestinal crypt populations dedifferentiate to provide new ISCs, but the transcriptional and signaling trajectories that guide this process are unclear, and a large body of work suggests that quiescent "reserve" ISCs contribute to regeneration. By timing the interval between LGR5+ lineage tracing and lethal injury, we show that ISC regeneration is explained nearly completely by dedifferentiation, with contributions from absorptive and secretory progenitors. The ISC-restricted transcription factor ASCL2 confers measurable competitive advantage to resting ISCs and is essential to restore the ISC compartment. Regenerating cells re-express Ascl2 days before Lgr5, and single-cell RNA sequencing (scRNA-seq) analyses reveal transcriptional paths underlying dedifferentiation. ASCL2 target genes include the interleukin-11 (IL-11) receptor Il11ra1, and recombinant IL-11 enhances crypt cell regenerative potential. These findings reveal cell dedifferentiation as the principal means for ISC restoration and highlight an ASCL2-regulated signal that enables this adaptive response.
Keywords: facultative stem cells; reserve stem cells; stem cell dedifferentiation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests H.C. is an inventor on patents related to intestinal organoids (full disclosure at https://www.uu.nl/staff/JCClevers/). The other authors declare no competing interests.
Figures


Comment in
-
Reserving the Right to Change the Intestinal Stem Cell Model.Cell Stem Cell. 2020 Mar 5;26(3):301-302. doi: 10.1016/j.stem.2020.02.003. Cell Stem Cell. 2020. PMID: 32142658 Free PMC article.
References
-
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. - PubMed
-
- Bamba S, Andoh A, Yasui H, Makino J, Kim S, and Fujiyama Y (2003). Regulation of IL-11 expression in intestinal myofibroblasts: role of c-Jun AP-1- and MAPK-dependent pathways. Am J Physiol Gastrointest Liver Physiol 285, G529–238. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

