Animal, Fungi, and Plant Genome Sequences Harbor Different Non-Canonical Splice Sites
- PMID: 32085510
- PMCID: PMC7072748
- DOI: 10.3390/cells9020458
Animal, Fungi, and Plant Genome Sequences Harbor Different Non-Canonical Splice Sites
Abstract
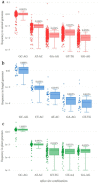
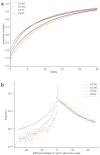
Most protein-encoding genes in eukaryotes contain introns, which are interwoven with exons. Introns need to be removed from initial transcripts in order to generate the final messenger RNA (mRNA), which can be translated into an amino acid sequence. Precise excision of introns by the spliceosome requires conserved dinucleotides, which mark the splice sites. However, there are variations of the highly conserved combination of GT at the 5' end and AG at the 3' end of an intron in the genome. GC-AG and AT-AC are two major non-canonical splice site combinations, which have been known for years. Recently, various minor non-canonical splice site combinations were detected with numerous dinucleotide permutations. Here, we expand systematic investigations of non-canonical splice site combinations in plants across eukaryotes by analyzing fungal and animal genome sequences. Comparisons of splice site combinations between these three kingdoms revealed several differences, such as an apparently increased CT-AC frequency in fungal genome sequences. Canonical GT-AG splice site combinations in antisense transcripts are a likely explanation for this observation, thus indicating annotation errors. In addition, high numbers of GA-AG splice site combinations were observed in Eurytemoraaffinis and Oikopleuradioica. A variant in one U1 small nuclear RNA (snRNA) isoform might allow the recognition of GA as a 5' splice site. In depth investigation of splice site usage based on RNA-Seq read mappings indicates a generally higher flexibility of the 3' splice site compared to the 5' splice site across animals, fungi, and plants.
Keywords: RNA-Seq; gene structure; introns; mRNA processing; sequence conservation; splice site analysis pipeline; spliceosome; splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genome-wide analyses supported by RNA-Seq reveal non-canonical splice sites in plant genomes.BMC Genomics. 2018 Dec 29;19(1):980. doi: 10.1186/s12864-018-5360-z. BMC Genomics. 2018. PMID: 30594132 Free PMC article.
-
Analysis of canonical and non-canonical splice sites in mammalian genomes.Nucleic Acids Res. 2000 Nov 1;28(21):4364-75. doi: 10.1093/nar/28.21.4364. Nucleic Acids Res. 2000. PMID: 11058137 Free PMC article.
-
Incorporation of splice site probability models for non-canonical introns improves gene structure prediction in plants.Bioinformatics. 2005 Nov 1;21 Suppl 3:iii20-30. doi: 10.1093/bioinformatics/bti1205. Bioinformatics. 2005. PMID: 16306388
-
Splice site requirements and switches in plants.Curr Top Microbiol Immunol. 2008;326:39-59. doi: 10.1007/978-3-540-76776-3_3. Curr Top Microbiol Immunol. 2008. PMID: 18630746 Review.
-
A novel role of U1 snRNP: Splice site selection from a distance.Biochim Biophys Acta Gene Regul Mech. 2019 Jun;1862(6):634-642. doi: 10.1016/j.bbagrm.2019.04.004. Epub 2019 Apr 28. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31042550 Free PMC article. Review.
Cited by
-
The reuse of public datasets in the life sciences: potential risks and rewards.PeerJ. 2020 Sep 22;8:e9954. doi: 10.7717/peerj.9954. eCollection 2020. PeerJ. 2020. PMID: 33024631 Free PMC article.
-
A rare non-canonical splice site in Trema orientalis SYMRK does not affect its dual symbiotic functioning in endomycorrhiza and rhizobium nodulation.BMC Plant Biol. 2023 Nov 24;23(1):587. doi: 10.1186/s12870-023-04594-0. BMC Plant Biol. 2023. PMID: 37996841 Free PMC article.
-
Beyond mass spectrometry, the next step in proteomics.Sci Adv. 2020 Jan 10;6(2):eaax8978. doi: 10.1126/sciadv.aax8978. eCollection 2020 Jan. Sci Adv. 2020. PMID: 31950079 Free PMC article. Review.
-
Quantitative and qualitative plant-pathogen interactions call upon similar pathogenicity genes with a spectrum of effects.Front Plant Sci. 2023 May 10;14:1128546. doi: 10.3389/fpls.2023.1128546. eCollection 2023. Front Plant Sci. 2023. PMID: 37235026 Free PMC article.
-
Where the minor things are: a pan-eukaryotic survey suggests neutral processes may explain much of minor intron evolution.Nucleic Acids Res. 2023 Nov 10;51(20):10884-10908. doi: 10.1093/nar/gkad797. Nucleic Acids Res. 2023. PMID: 37819006 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

