The ER Unfolded Protein Response Effector, ATF6, Reduces Cardiac Fibrosis and Decreases Activation of Cardiac Fibroblasts
- PMID: 32085622
- PMCID: PMC7073073
- DOI: 10.3390/ijms21041373
The ER Unfolded Protein Response Effector, ATF6, Reduces Cardiac Fibrosis and Decreases Activation of Cardiac Fibroblasts
Abstract
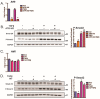
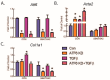
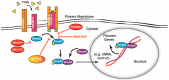
Activating transcription factor-6 α (ATF6) is one of the three main sensors and effectors of the endoplasmic reticulum (ER) stress response and, as such, it is critical for protecting the heart and other tissues from a variety of environmental insults and disease states. In the heart, ATF6 has been shown to protect cardiac myocytes. However, its roles in other cell types in the heart are unknown. Here we show that ATF6 decreases the activation of cardiac fibroblasts in response to the cytokine, transforming growth factor β (TGFβ), which can induce fibroblast trans-differentiation into a myofibroblast phenotype through signaling via the TGFβ-Smad pathway. ATF6 activation suppressed fibroblast contraction and the induction of α smooth muscle actin (αSMA). Conversely, fibroblasts were hyperactivated when ATF6 was silenced or deleted. ATF6 thus represents a novel inhibitor of the TGFβ-Smad axis of cardiac fibroblast activation.
Keywords: ATF6; ER stress; Smad; TGFβ; UPR; cardiac fibroblast; cardiac fibrosis; endoplasmic reticulum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
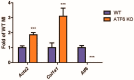




References
-
- Jurkin J., Henkel T., Nielsen A.F., Minnich M., Popow J., Kaufmann T., Heindl K., Hoffmann T., Busslinger M., Martinez J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 2014;33:2922–2936. doi: 10.15252/embj.201490332. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

