Molecular detection and phylogenetic analysis of lumpy skin disease virus from outbreaks in Uganda 2017-2018
- PMID: 32085763
- PMCID: PMC7035724
- DOI: 10.1186/s12917-020-02288-5
Molecular detection and phylogenetic analysis of lumpy skin disease virus from outbreaks in Uganda 2017-2018
Abstract
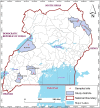
Background: Lumpy skin disease (LSD) is an infectious viral disease of cattle caused by a Capripoxvirus. LSD has substantial economic implications, with infection resulting in permanent damage to the skin of affected animals which lowers their commercial value. In Uganda, LSD is endemic and cases of the disease are frequently reported to government authorities. This study was undertaken to molecularly characterize lumpy skin disease virus (LSDV) strains that have been circulating in Uganda between 2017 and 2018. Secondly, the study aimed to determine the phylogenetic relatedness of Ugandan LSDV sequences with published sequences, available in GenBank.
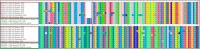
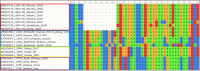
Results: A total of 7 blood samples and 16 skin nodule biopsies were screened for LSDV using PCR to confirm presence of LSDV nucleic acids. PCR positive samples were then characterised by amplifying the GPCR gene. These amplified genes were sequenced and phylogenetic trees were constructed. Out of the 23 samples analysed, 15 were positive for LSDV by PCR (65.2%). The LSDV GPCR sequences analysed contained the unique signatures of LSDV (A11, T12, T34, S99, and P199) which further confirmed their identity. Sequence comparison with vaccine strains revealed a 12 bp deletion unique to Ugandan outbreak strains. Phylogenetic analysis indicated that the LSDV sequences from this study clustered closely with sequences from neighboring East African countries and with LSDV strains from recent outbreaks in Europe. It was noted that the sequence diversity amongst LSDV strains from Africa was higher than diversity from Eurasia.
Conclusion: The LSDV strains circulating in Uganda were closely related with sequences from neighboring African countries and from Eurasia. Comparison of the GPCR gene showed that outbreak strains differed from vaccine strains. This information is necessary to understand LSDV molecular epidemiology and to contribute knowledge towards the development of control strategies by the Government of Uganda.
Keywords: GPCR; Lumpy skin disease; Molecular detection; Phylogenetic analysis; Uganda.
Conflict of interest statement
The authors of this paper do not have any financial or personal relationship with other people or organisations that could inappropriately influence or bias the content of the paper. The authors therefore declare that they have no competing interests in the publication of this paper.
Figures


References
-
- Fauquet C., Mayo M.A., Maniloff J. DU& BLA. Virus Taxonomy: VIII Report of theInternational Committee onTaxonomy ofViruses. Elsevier Acad. Press. San Diego, California, USA London, UK. 2005;
-
- Davies FG. Lumpy skin disease of cattle: a growing problem in Africa and the near east. World Anim Rev 1991;68:37–42. Available from: http://www.fao.org/ag/aGa/agap/frg/feedback/war/u4900b/u4900b0d.htm
-
- OIE. Lumpy Skin Disease. OIE Terr. Man. [Internet]. 2017. p. 56. Available from: https://www.bertelsmann-stiftung.de/fileadmin/files/BSt/Publikationen/Gr... society and inequalities%28lsero%29.pdf%0Ahttps://www.quora.com/What-is-the
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

