Can rumen bacteria communicate to each other?
- PMID: 32085816
- PMCID: PMC7035670
- DOI: 10.1186/s40168-020-00796-y
Can rumen bacteria communicate to each other?
Abstract
Background: The rumen contains a myriad of microbes whose primary role is to degrade and ferment dietary nutrients, which then provide the host with energy and nutrients. Rumen microbes commonly attach to ingested plant materials and form biofilms for effective plant degradation. Quorum sensing (QS) is a well-recognised form of bacterial communication in most biofilm communities, with homoserine lactone (AHL)-based QS commonly being used by Gram-negative bacteria alone and AI-2 Lux-based QS communication being used to communicate across Gram-negative and Gram-positive bacteria. However, bacterial cell to cell communication in the rumen is poorly understood. In this study, rumen bacterial genomes from the Hungate collection and Genbank were prospected for QS-related genes. To check that the discovered QS genes are actually expressed in the rumen, we investigated expression levels in rumen metatranscriptome datasets.
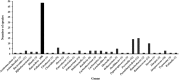
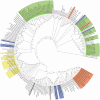
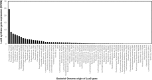
Results: A total of 448 rumen bacterial genomes from the Hungate collection and Genbank, comprised of 311 Gram-positive, 136 Gram-negative and 1 Gram stain variable bacterium, were analysed. Abundance and distribution of AHL and AI-2 signalling genes showed that only one species (Citrobacter sp. NLAE-zl-C269) of a Gram-negative bacteria appeared to possess an AHL synthase gene, while the Lux-based genes (AI-2 QS) were identified in both Gram-positive and Gram-positive bacteria (191 genomes representing 38.2% of total genomes). Of these 192 genomes, 139 are from Gram-positive bactreetteria and 53 from Gram-negative bacteria. We also found that the genera Butyrivibrio, Prevotella, Ruminococcus and Pseudobutyrivibrio, which are well known as the most abundant bacterial genera in the rumen, possessed the most lux-based AI-2 QS genes. Gene expression levels within the metatranscriptome dataset showed that Prevotella, in particular, expressed high levels of LuxS synthase suggesting that this genus plays an important role in QS within the rumen.
Conclusion: This is the most comprehensive study of QS in the rumen microbiome to date. This study shows that AI-2-based QS is rife in the rumen. These results allow a greater understanding on plant-microbe interactions in the rumen.
Keywords: AI-2; Acyl-homoserine lactone; Bacteria; LuxS; Quorum sensing; Rumen.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

