Molecular dynamics simulations suggest stabilizing mutations in a de novo designed α/β protein
- PMID: 32086513
- PMCID: PMC7052480
- DOI: 10.1093/protein/gzaa005
Molecular dynamics simulations suggest stabilizing mutations in a de novo designed α/β protein
Abstract
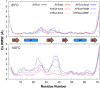
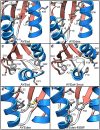
Designing functional proteins that can withstand extreme heat is beneficial for industrial and protein therapeutic applications. Thus, elucidating the atomic-level determinants of thermostability is a major interest for rational protein design. To that end, we compared the structure and dynamics of a set of previously designed, thermostable proteins based on the activation domain of human procarboxypeptidase A2 (AYEwt). The mutations in these designed proteins were intended to increase hydrophobic core packing and inter-secondary-structure interactions. To evaluate whether these design strategies were successfully deployed, we performed all-atom, explicit-solvent molecular dynamics (MD) simulations of AYEwt and three designed variants at both 25 and 100°C. Our MD simulations agreed with the relative experimental stabilities of the designs based on their secondary structure content, Cα root-mean-square deviation/fluctuation, and buried-residue solvent accessible surface area. Using a contact analysis, we found that the designs stabilize inter-secondary structure interactions and buried hydrophobic surface area, as intended. Based on our analysis, we designed three additional variants to test the role of helix stabilization, core packing, and a Phe → Met mutation on thermostability. We performed the additional MD simulations and analysis on these variants, and these data supported our predictions.
Keywords: molecular dynamics; protein design; protein thermostability.
© The Author(s) 2020. Published by Oxford University Press.
Figures




Similar articles
-
The stability and dynamics of computationally designed proteins.Protein Eng Des Sel. 2022 Feb 17;35:gzac001. doi: 10.1093/protein/gzac001. Protein Eng Des Sel. 2022. PMID: 35174855 Free PMC article. Review.
-
A Dynamic Hydrophobic Core and Surface Salt Bridges Thermostabilize a Designed Three-Helix Bundle.Biophys J. 2019 Feb 19;116(4):621-632. doi: 10.1016/j.bpj.2019.01.012. Epub 2019 Jan 12. Biophys J. 2019. PMID: 30704856 Free PMC article.
-
Rational Design of Thermostable Carbonic Anhydrase Mutants Using Molecular Dynamics Simulations.J Phys Chem B. 2018 Sep 13;122(36):8526-8536. doi: 10.1021/acs.jpcb.8b05926. Epub 2018 Aug 29. J Phys Chem B. 2018. PMID: 30114369
-
Lys-Arg mutation improved the thermostability of Bacillus cereus neutral protease through increased residue interactions.World J Microbiol Biotechnol. 2019 Oct 31;35(11):173. doi: 10.1007/s11274-019-2751-5. World J Microbiol Biotechnol. 2019. PMID: 31673794
-
Analysis of molecular dynamics simulations of 10-residue peptide, chignolin, using statistical mechanics: Relaxation mode analysis and three-dimensional reference interaction site model theory.Biophys Physicobiol. 2019 Nov 29;16:407-429. doi: 10.2142/biophysico.16.0_407. eCollection 2019. Biophys Physicobiol. 2019. PMID: 31984194 Free PMC article. Review.
Cited by
-
De novo design of drug-binding proteins with predictable binding energy and specificity.Science. 2024 Apr 5;384(6691):106-112. doi: 10.1126/science.adl5364. Epub 2024 Apr 4. Science. 2024. PMID: 38574125 Free PMC article.
-
KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability.Int J Mol Sci. 2021 May 20;22(10):5408. doi: 10.3390/ijms22105408. Int J Mol Sci. 2021. PMID: 34065616 Free PMC article.
-
The stability and dynamics of computationally designed proteins.Protein Eng Des Sel. 2022 Feb 17;35:gzac001. doi: 10.1093/protein/gzac001. Protein Eng Des Sel. 2022. PMID: 35174855 Free PMC article. Review.
-
dUTP pyrophosphatases from hyperthermophilic eubacterium and archaeon: Structural and functional examinations on the suitability for PCR application.Protein Sci. 2024 Nov;33(11):e5185. doi: 10.1002/pro.5185. Protein Sci. 2024. PMID: 39440877
-
De novo design of drug-binding proteins with predictable binding energy and specificity.bioRxiv [Preprint]. 2023 Dec 23:2023.12.23.573178. doi: 10.1101/2023.12.23.573178. bioRxiv. 2023. Update in: Science. 2024 Apr 5;384(6691):106-112. doi: 10.1126/science.adl5364. PMID: 38187746 Free PMC article. Updated. Preprint.
References
-
- Beck D.A.C., McCully M.E., Alonso D.O.V., Daggett V. (2000) In lucem molecular mechanics (in lucem molecular mechanics (ilmm) 2000–2020). University of Washington, Seattle.
-
- Becktel W.J. and Schellman J.A. (1987) Biopolymers, 26, 1859–1877. - PubMed
-
- Beglov D. and Roux B. (1998) J. Chem. Phys., 100, 9050.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

