FIGLA, LHX8 and SOHLH1 transcription factor networks regulate mouse oocyte growth and differentiation
- PMID: 32086523
- PMCID: PMC7144910
- DOI: 10.1093/nar/gkaa101
FIGLA, LHX8 and SOHLH1 transcription factor networks regulate mouse oocyte growth and differentiation
Abstract
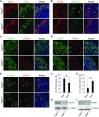
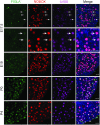
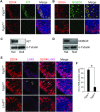
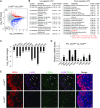
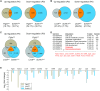
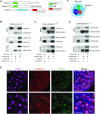
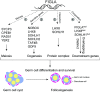
Germ-cell transcription factors control gene networks that regulate oocyte differentiation and primordial follicle formation during early, postnatal mouse oogenesis. Taking advantage of gene-edited mice lacking transcription factors expressed in female germ cells, we analyzed global gene expression profiles in perinatal ovaries from wildtype, FiglaNull, Lhx8Null and Sohlh1Null mice. Figla deficiency dysregulates expression of meiosis-related genes (e.g. Sycp3, Rad51, Ybx2) and a variety of genes (e.g. Nobox, Lhx8, Taf4b, Sohlh1, Sohlh2, Gdf9) associated with oocyte growth and differentiation. The absence of FIGLA significantly impedes meiotic progression, causes DNA damage and results in oocyte apoptosis. Moreover, we find that FIGLA and other transcriptional regulator proteins (e.g. NOBOX, LHX8, SOHLH1, SOHLH2) are co-expressed in the same subset of germ cells in perinatal ovaries and Figla ablation dramatically disrupts KIT, NOBOX, LHX8, SOHLH1 and SOHLH2 abundance. In addition, not only do FIGLA, LHX8 and SOHLH1 cross-regulate each other, they also cooperate by direct interaction with each during early oocyte development and share downstream gene targets. Thus, our findings substantiate a major role for FIGLA, LHX8 and SOHLH1 as multifunctional regulators of networks necessary for oocyte maintenance and differentiation during early folliculogenesis.
Published by Oxford University Press on behalf of Nucleic Acids Research 2020.
Figures


Similar articles
-
Transcription factors SOHLH1 and SOHLH2 coordinate oocyte differentiation without affecting meiosis I.J Clin Invest. 2017 Jun 1;127(6):2106-2117. doi: 10.1172/JCI90281. Epub 2017 May 15. J Clin Invest. 2017. PMID: 28504655 Free PMC article.
-
Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8.Proc Natl Acad Sci U S A. 2006 May 23;103(21):8090-5. doi: 10.1073/pnas.0601083103. Epub 2006 May 11. Proc Natl Acad Sci U S A. 2006. PMID: 16690745 Free PMC article.
-
Lhx8 regulates primordial follicle activation and postnatal folliculogenesis.BMC Biol. 2015 Jun 16;13:39. doi: 10.1186/s12915-015-0151-3. BMC Biol. 2015. PMID: 26076587 Free PMC article.
-
Oogenesis: transcriptional regulators and mouse models.Mol Cell Endocrinol. 2012 Jun 5;356(1-2):31-9. doi: 10.1016/j.mce.2011.07.049. Epub 2011 Aug 12. Mol Cell Endocrinol. 2012. PMID: 21856374 Review.
-
Transcriptional regulation of early oogenesis: in search of masters.Hum Reprod Update. 2006 Jan-Feb;12(1):65-76. doi: 10.1093/humupd/dmi033. Epub 2005 Sep 2. Hum Reprod Update. 2006. PMID: 16143663 Review.
Cited by
-
The transcriptome-wide N6-methyladenosine (m6A) map profiling reveals the regulatory role of m6A in the yak ovary.BMC Genomics. 2022 May 11;23(1):358. doi: 10.1186/s12864-022-08585-7. BMC Genomics. 2022. PMID: 35538402 Free PMC article.
-
LSM14B controls oocyte mRNA storage and stability to ensure female fertility.Cell Mol Life Sci. 2023 Aug 14;80(9):247. doi: 10.1007/s00018-023-04898-2. Cell Mol Life Sci. 2023. PMID: 37578641 Free PMC article.
-
RAD51 and Infertility: A Review and Case-Control Study.Biochem Genet. 2024 Apr;62(2):1216-1230. doi: 10.1007/s10528-023-10469-8. Epub 2023 Aug 10. Biochem Genet. 2024. PMID: 37563467 Review.
-
Efficacy of Sailuotong on neurovascular unit in amyloid precursor protein/presenilin-1 transgenic mice with Alzheimer's disease.J Tradit Chin Med. 2024 Apr;44(2):289-302. doi: 10.19852/j.cnki.jtcm.20240203.007. J Tradit Chin Med. 2024. PMID: 38504535 Free PMC article.
-
A matter of new life and cell death: programmed cell death in the mammalian ovary.J Biomed Sci. 2024 Mar 20;31(1):31. doi: 10.1186/s12929-024-01017-6. J Biomed Sci. 2024. PMID: 38509545 Free PMC article. Review.
References
-
- Pepling M.E., Spradling A.C.. Female mouse germ cells form synchronously dividing cysts. Development. 1998; 125:3323–3328. - PubMed
-
- Pepling M.E., Spradling A.C.. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev. Biol. 2001; 234:339–351. - PubMed
-
- Pepling M.E. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis. 2006; 44:622–632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

