Heterochromatin loss as a determinant of progerin-induced DNA damage in Hutchinson-Gilford Progeria
- PMID: 32087607
- PMCID: PMC7059134
- DOI: 10.1111/acel.13108
Heterochromatin loss as a determinant of progerin-induced DNA damage in Hutchinson-Gilford Progeria
Abstract
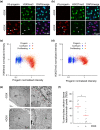
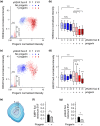
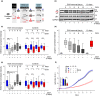
Hutchinson-Gilford progeria is a premature aging syndrome caused by a truncated form of lamin A called progerin. Progerin expression results in a variety of cellular defects including heterochromatin loss, DNA damage, impaired proliferation and premature senescence. It remains unclear how these different progerin-induced phenotypes are temporally and mechanistically linked. To address these questions, we use a doxycycline-inducible system to restrict progerin expression to different stages of the cell cycle. We find that progerin expression leads to rapid and widespread loss of heterochromatin in G1-arrested cells, without causing DNA damage. In contrast, progerin triggers DNA damage exclusively during late stages of DNA replication, when heterochromatin is normally replicated, and preferentially in cells that have lost heterochromatin. Importantly, removal of progerin from G1-arrested cells restores heterochromatin levels and results in no permanent proliferative impediment. Taken together, these results delineate the chain of events that starts with progerin expression and ultimately results in premature senescence. Moreover, they provide a proof of principle that removal of progerin from quiescent cells restores heterochromatin levels and their proliferative capacity to normal levels.
Keywords: DNA damage; HGPS; heterochromatin; lamin A; progerin; senescence.
© 2020 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
Authors declare that there is no conflict of interest.
Figures


References
-
- Arnoult, N. , Schluth‐Bolard, C. , Letessier, A. , Drascovic, I. , Bouarich‐Bourimi, R. , Campisi, J. , … Londoño‐Vallejo, A. (2010). Replication timing of human telomeres is chromosome arm‐specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genetics, 6, e1000920 10.1371/journal.pgen.1000920 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

