The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region
- PMID: 32087618
- PMCID: PMC7060142
- DOI: 10.1111/mpp.12894
The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region
Abstract
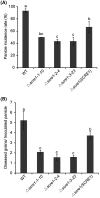
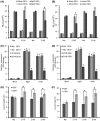
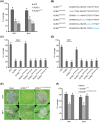
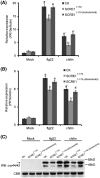
The biotrophic fungal pathogen Ustilaginoidea virens causes rice false smut, a newly emerging plant disease that has become epidemic worldwide in recent years. The U. virens genome encodes many putative effector proteins that, based on the study of other pathosystems, could play an essential role in fungal virulence. However, few studies have been reported on virulence functions of individual U. virens effectors. Here, we report our identification and characterization of the secreted cysteine-rich protein SCRE1, which is an essential virulence effector in U. virens. When SCRE1 was heterologously expressed in Magnaporthe oryzae, the protein was secreted and translocated into plant cells during infection. SCRE1 suppresses the immunity-associated hypersensitive response in the nonhost plant Nicotiana benthamiana. Induced expression of SCRE1 in rice also inhibits pattern-triggered immunity and enhances disease susceptibility to rice bacterial and fungal pathogens. The immunosuppressive activity is localized to a small peptide region that contains an important 'cysteine-proline-alanine-arginine-serine' motif. Furthermore, the scre1 knockout mutant generated using the CRISPR/Cas9 system is attenuated in U. virens virulence to rice, which is greatly complemented by the full-length SCRE1 gene. Collectively, this study indicates that the effector SCRE1 is able to inhibit host immunity and is required for full virulence of U. virens.
Keywords: Ustilaginoidea virens; SCRE1; fungal effector; immunosuppressive peptide; rice immunity.
© 2019 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures



References
-
- Ai, H. , Shaner, N.C. , Cheng, Z. and Tsien, R.Y . and Campbell, R.E. (2007) Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry, 46, 5904–5910. - PubMed
-
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. - PubMed
-
- An, G. , Ebert, P.R. , Mitra, A. and Ha, S.B. (1988) Binary Vectors In: Plant Molecular Biology Manual (Gelvin S.B. and Schilperoort R.A., eds), pp. A3, 1–19. Dordrecht, Netherlands: Kluwer Academic Publishers.
-
- Aoyama, T. and Chua, N.H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

