Computational MHC-I epitope predictor identifies 95% of experimentally mapped HIV-1 clade A and D epitopes in a Ugandan cohort
- PMID: 32087680
- PMCID: PMC7036183
- DOI: 10.1186/s12879-020-4876-4
Computational MHC-I epitope predictor identifies 95% of experimentally mapped HIV-1 clade A and D epitopes in a Ugandan cohort
Abstract
Background: Identifying immunogens that induce HIV-1-specific immune responses is a lengthy process that can benefit from computational methods, which predict T-cell epitopes for various HLA types.
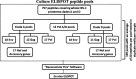
Methods: We tested the performance of the NetMHCpan4.0 computational neural network in re-identifying 93 T-cell epitopes that had been previously independently mapped using the whole proteome IFN-γ ELISPOT assays in 6 HLA class I typed Ugandan individuals infected with HIV-1 subtypes A1 and D. To provide a benchmark we compared the predictions for NetMHCpan4.0 to MHCflurry1.2.0 and NetCTL1.2.
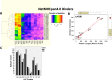
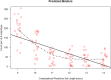
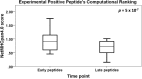
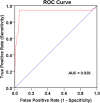
Results: NetMHCpan4.0 performed best correctly predicting 88 of the 93 experimentally mapped epitopes for a set length of 9-mer and matched HLA class I alleles. Receiver Operator Characteristic (ROC) analysis gave an area under the curve (AUC) of 0.928. Setting NetMHCpan4.0 to predict 11-14mer length did not improve the prediction (37-79 of 93 peptides) with an inverse correlation between the number of predictions and length set. Late time point peptides were significantly stronger binders than early peptides (Wilcoxon signed rank test: p = 0.0000005). MHCflurry1.2.0 similarly predicted all but 2 of the peptides that NetMHCpan4.0 predicted and NetCTL1.2 predicted only 14 of the 93 experimental peptides.
Conclusion: NetMHCpan4.0 class I epitope predictions covered 95% of the epitope responses identified in six HIV-1 infected individuals, and would have reduced the number of experimental confirmatory tests by > 80%. Algorithmic epitope prediction in conjunction with HLA allele frequency information can cost-effectively assist immunogen design through minimizing the experimental effort.
Keywords: Artificial neural network; Epitope mapping; HIV-1; In-silico; MHCflurry1.2.0 and NetCTL1.2; NetMHCpan4.0.; T-cell.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Azizi A, Anderson DE, Torres JV, Ogrel A, Ghorbani M, Soare C, Sandstrom P, Fournier J, Diaz-Mitoma F. Induction of broad cross-subtype-specific HIV-1 immune responses by a novel multivalent HIV-1 peptide vaccine in cynomolgus macaques. J Immunol. 2008;180(4):2174–2186. doi: 10.4049/jimmunol.180.4.2174. - DOI - PubMed
-
- Baden LR, Walsh SR, Seaman MS, Cohen YZ, Johnson JA, Licona JH, Filter RD, Kleinjan JA, Gothing JA, Jennings J, et al. First-in-human randomized controlled trial of mosaic HIV-1 Immunogens delivered via a modified Vaccinia Ankara vector. J Infect Dis. 2018;218(4):633–644. doi: 10.1093/infdis/jiy212. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- MC_UU_00027/1/MRC_/Medical Research Council/United Kingdom
- TA_05_40200_40203/European and Developing Countries Clinical Trials Partnership
- WT078927MA/UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement, the Wellcome Trust
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

