Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer
- PMID: 32091413
- PMCID: PMC7108892
- DOI: 10.1172/JCI131041
Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer
Abstract
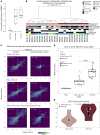
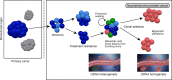
Loss of androgen receptor (AR) signaling dependence occurs in approximately 15%-20% of advanced treatment-resistant prostate cancers, and this may manifest clinically as transformation from a prostate adenocarcinoma histology to a castration-resistant neuroendocrine prostate cancer (CRPC-NE). The diagnosis of CRPC-NE currently relies on a metastatic tumor biopsy, which is invasive for patients and sometimes challenging to diagnose due to morphologic heterogeneity. By studying whole-exome sequencing and whole-genome bisulfite sequencing of cell free DNA (cfDNA) and of matched metastatic tumor biopsies from patients with metastatic prostate adenocarcinoma and CRPC-NE, we identified CRPC-NE features detectable in the circulation. Overall, there was markedly higher concordance between cfDNA and biopsy tissue genomic alterations in patients with CRPC-NE compared with castration-resistant adenocarcinoma, supporting greater intraindividual genomic consistency across metastases. Allele-specific copy number and serial sampling analyses allowed for the detection and tracking of clonal and subclonal tumor cell populations. cfDNA methylation was indicative of circulating tumor content fraction, reflective of methylation patterns observed in biopsy tissues, and was capable of detecting CRPC-NE-associated epigenetic changes (e.g., hypermethylation of ASXL3 and SPDEF; hypomethylation of INSM1 and CDH2). A targeted set combining genomic (TP53, RB1, CYLD, AR) and epigenomic (hypo- and hypermethylation of 20 differential sites) alterations applied to ctDNA was capable of identifying patients with CRPC-NE.
Keywords: Oncology; Prostate cancer.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

