WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line
- PMID: 32093151
- PMCID: PMC7072831
- DOI: 10.3390/cells9020485
WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line
Erratum in
-
Correction: Alves et al. WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line. Cells 2020, 9, 485.Cells. 2024 Jul 19;13(14):1219. doi: 10.3390/cells13141219. Cells. 2024. PMID: 39056814 Free PMC article.
Abstract
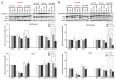
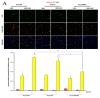
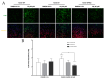
Autophagy is a cell-survival pathway with dual role in tumorigenesis, promoting either tumor survival or tumor death. WNK2 gene, a member of the WNK (with no lysine (K)) subfamily, acts as a tumor suppressor gene in gliomas, regulating cell migration and invasion; however, its role in autophagy process is poorly explored. The WNK2-methylated human glioblastoma cell line A172 WT (wild type) was compared to transfected clones A172 EV (empty vector), and A172 WNK2 (WNK2 overexpression) for the evaluation of autophagy using an inhibitor (bafilomycin A1-baf A1) and an inducer (everolimus) of autophagic flux. Western blot and immunofluorescence approaches were used to monitor autophagic markers, LC3A/B and SQSTM1/p62. A172 WNK2 cells presented a significant decrease in LC3B and p62 protein levels, and in LC3A/B ratio when compared with control cells, after treatment with baf A1 + everolimus, suggesting that WNK2 overexpression inhibits the autophagic flux in gliomas. The mTOR pathway was also evaluated under the same conditions, and the observed results suggest that the inhibition of autophagy mediated by WNK2 occurs through a mTOR-independent pathway. In conclusion, the evaluation of the autophagic process demonstrated that WNK2 inhibits the autophagic flux in glioblastoma cell line.
Keywords: WNK2; autophagic flux; autophagy; glioblastoma cell line; inhibition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

