Unexpected cell type-dependent effects of autophagy on polyglutamine aggregation revealed by natural genetic variation in C. elegans
- PMID: 32093691
- PMCID: PMC7038566
- DOI: 10.1186/s12915-020-0750-5
Unexpected cell type-dependent effects of autophagy on polyglutamine aggregation revealed by natural genetic variation in C. elegans
Abstract
Background: Monogenic protein aggregation diseases, in addition to cell selectivity, exhibit clinical variation in the age of onset and progression, driven in part by inter-individual genetic variation. While natural genetic variants may pinpoint plastic networks amenable to intervention, the mechanisms by which they impact individual susceptibility to proteotoxicity are still largely unknown.
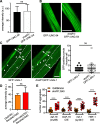
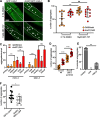
Results: We have previously shown that natural variation modifies polyglutamine (polyQ) aggregation phenotypes in C. elegans muscle cells. Here, we find that a genomic locus from C. elegans wild isolate DR1350 causes two genetically separable aggregation phenotypes, without changing the basal activity of muscle proteostasis pathways known to affect polyQ aggregation. We find that the increased aggregation phenotype was due to regulatory variants in the gene encoding a conserved autophagy protein ATG-5. The atg-5 gene itself conferred dosage-dependent enhancement of aggregation, with the DR1350-derived allele behaving as hypermorph. Surprisingly, increased aggregation in animals carrying the modifier locus was accompanied by enhanced autophagy activation in response to activating treatment. Because autophagy is expected to clear, not increase, protein aggregates, we activated autophagy in three different polyQ models and found a striking tissue-dependent effect: activation of autophagy decreased polyQ aggregation in neurons and intestine, but increased it in the muscle cells.
Conclusions: Our data show that cryptic natural variants in genes encoding proteostasis components, although not causing detectable phenotypes in wild-type individuals, can have profound effects on aggregation-prone proteins. Clinical applications of autophagy activators for aggregation diseases may need to consider the unexpected divergent effects of autophagy in different cell types.
Keywords: Autophagy; Cryptic variation; Natural genetic variation; Polyglutamine; Protein aggregation; Proteostasis; Regulatory variation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

