Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD
- PMID: 32093728
- PMCID: PMC7041170
- DOI: 10.1186/s13024-020-00365-9
Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD
Abstract
Background: The C9ORF72 hexanucleotide repeat expansion is the most common known genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), two fatal age-related neurodegenerative diseases. The C9ORF72 expansion encodes five dipeptide repeat proteins (DPRs) that are produced through a non-canonical translation mechanism. Among the DPRs, proline-arginine (PR), glycine-arginine (GR), and glycine-alanine (GA) are the most neurotoxic and increase the frequency of DNA double strand breaks (DSBs). While the accumulation of these genotoxic lesions is increasingly recognized as a feature of disease, the mechanism(s) of DPR-mediated DNA damage are ill-defined and the effect of DPRs on the efficiency of each DNA DSB repair pathways has not been previously evaluated.
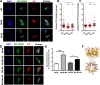
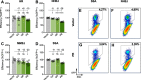
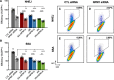
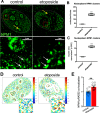
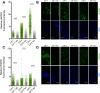
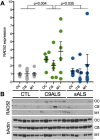
Methods and results: Using DNA DSB repair assays, we evaluated the efficiency of specific repair pathways, and found that PR, GR and GA decrease the efficiency of non-homologous end joining (NHEJ), single strand annealing (SSA), and microhomology-mediated end joining (MMEJ), but not homologous recombination (HR). We found that PR inhibits DNA DSB repair, in part, by binding to the nucleolar protein nucleophosmin (NPM1). Depletion of NPM1 inhibited NHEJ and SSA, suggesting that NPM1 loss-of-function in PR expressing cells leads to impediments of both non-homologous and homology-directed DNA DSB repair pathways. By deleting NPM1 sub-cellular localization signals, we found that PR binds NPM1 regardless of the cellular compartment to which NPM1 was directed. Deletion of the NPM1 acidic loop motif, known to engage other arginine-rich proteins, abrogated PR and NPM1 binding. Using confocal and super-resolution immunofluorescence microscopy, we found that levels of RAD52, a component of the SSA repair machinery, were significantly increased iPSC neurons relative to isogenic controls in which the C9ORF72 expansion had been deleted using CRISPR/Cas9 genome editing. Western analysis of post-mortem brain tissues confirmed that RAD52 immunoreactivity is significantly increased in C9ALS/FTD samples as compared to controls.
Conclusions: Collectively, we characterized the inhibitory effects of DPRs on key DNA DSB repair pathways, identified NPM1 as a facilitator of DNA repair that is inhibited by PR, and revealed deficits in homology-directed DNA DSB repair pathways as a novel feature of C9ORF72-related disease.
Keywords: Amyotrophic lateral sclerosis; CRISPR; DNA damage; DNA double strand break repair; Homology-directed repair; Induced pluripotent stem cells; RAD52; Single-strand annealing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

