Myosin heavy chain-embryonic regulates skeletal muscle differentiation during mammalian development
- PMID: 32094117
- PMCID: PMC7157585
- DOI: 10.1242/dev.184507
Myosin heavy chain-embryonic regulates skeletal muscle differentiation during mammalian development
Abstract
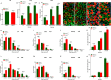
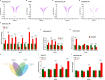
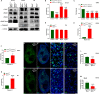
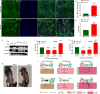
Myosin heavy chain-embryonic (MyHC-emb) is a skeletal muscle-specific contractile protein expressed during muscle development. Mutations in MYH3, the gene encoding MyHC-emb, lead to Freeman-Sheldon and Sheldon-Hall congenital contracture syndromes. Here, we characterize the role of MyHC-emb during mammalian development using targeted mouse alleles. Germline loss of MyHC-emb leads to neonatal and postnatal alterations in muscle fiber size, fiber number, fiber type and misregulation of genes involved in muscle differentiation. Deletion of Myh3 during embryonic myogenesis leads to the depletion of the myogenic progenitor cell pool and an increase in the myoblast pool, whereas fetal myogenesis-specific deletion of Myh3 causes the depletion of both myogenic progenitor and myoblast pools. We reveal that the non-cell-autonomous effect of MyHC-emb on myogenic progenitors and myoblasts is mediated by the fibroblast growth factor (FGF) signaling pathway, and exogenous FGF rescues the myogenic differentiation defects upon loss of MyHC-emb function in vitro Adult Myh3 null mice exhibit scoliosis, a characteristic phenotype exhibited by individuals with Freeman-Sheldon and Sheldon-Hall congenital contracture syndrome. Thus, we have identified MyHC-emb as a crucial myogenic regulator during development, performing dual cell-autonomous and non-cell-autonomous functions.This article has an associated 'The people behind the papers' interview.
Keywords: Development; FGF; Mice; Muscle progenitors; Myogenesis; Myosin heavy chain-embryonic; Signaling; Skeletal muscle.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

