Genomic Epidemiology of Complex, Multispecies, Plasmid-Borne blaKPC Carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014
- PMID: 32094139
- PMCID: PMC7179641
- DOI: 10.1128/AAC.02244-19
Genomic Epidemiology of Complex, Multispecies, Plasmid-Borne blaKPC Carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014
Abstract
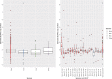
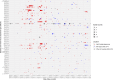
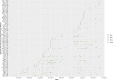
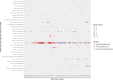
Carbapenem resistance in Enterobacterales is a public health threat. Klebsiella pneumoniae carbapenemase (encoded by alleles of the blaKPC family) is one of the most common transmissible carbapenem resistance mechanisms worldwide. The dissemination of blaKPC historically has been associated with distinct K. pneumoniae lineages (clonal group 258 [CG258]), a particular plasmid family (pKpQIL), and a composite transposon (Tn4401). In the United Kingdom, blaKPC has represented a large-scale, persistent management challenge for some hospitals, particularly in North West England. The dissemination of blaKPC has evolved to be polyclonal and polyspecies, but the genetic mechanisms underpinning this evolution have not been elucidated in detail; this study used short-read whole-genome sequencing of 604 blaKPC-positive isolates (Illumina) and long-read assembly (PacBio)/polishing (Illumina) of 21 isolates for characterization. We observed the dissemination of blaKPC (predominantly blaKPC-2; 573/604 [95%] isolates) across eight species and more than 100 known sequence types. Although there was some variation at the transposon level (mostly Tn4401a, 584/604 [97%] isolates; predominantly with ATTGA-ATTGA target site duplications, 465/604 [77%] isolates), blaKPC spread appears to have been supported by highly fluid, modular exchange of larger genetic segments among plasmid populations dominated by IncFIB (580/604 isolates), IncFII (545/604 isolates), and IncR (252/604 isolates) replicons. The subset of reconstructed plasmid sequences (21 isolates, 77 plasmids) also highlighted modular exchange among non-blaKPC and blaKPC plasmids and the common presence of multiple replicons within blaKPC plasmid structures (>60%). The substantial genomic plasticity observed has important implications for our understanding of the epidemiology of transmissible carbapenem resistance in Enterobacterales for the implementation of adequate surveillance approaches and for control.
Keywords: Enterobacterales; KPC; carbapenemase; genomic epidemiology; long-read sequencing; outbreak analysis; short-read sequencing; whole-genome sequencing.
Copyright © 2020 American Society for Microbiology.
Figures




References
-
- Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. 2015. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 59:1656–1668. doi:10.1128/AAC.04292-14. - DOI - PMC - PubMed
-
- Cerqueira GC, Earl AM, Ernst CM, Grad YH, Dekker JP, Feldgarden M, Chapman SB, Reis-Cunha JL, Shea TP, Young S, Zeng Q, Delaney ML, Kim D, Peterson EM, O'Brien TF, Ferraro MJ, Hooper DC, Huang SS, Kirby JE, Onderdonk AB, Birren BW, Hung DT, Cosimi LA, Wortman JR, Murphy CI, Hanage WP. 2017. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 114:1135–1140. doi:10.1073/pnas.1616248114. - DOI - PMC - PubMed
-
- Ruiz-Garbajosa P, Curiao T, Tato M, Gijón D, Pintado V, Valverde A, Baquero F, Morosini MI, Coque TM, Cantón R. 2013. Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother 68:2487–2492. doi:10.1093/jac/dkt237. - DOI - PubMed
-
- Martin J, Phan H, Findlay J, Stoesser N, Pankhurst L, Navickaite I, De Maio N, Eyre D, Toogood G, Orsi N, Kirby A, Young N, Turton J, Hill R, Hopkins K, Woodford N, Peto T, Walker A, Crook D, Wilcox M. 2017. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J Antimicrob Chemother 72:3025–3034. doi:10.1093/jac/dkx264. - DOI - PMC - PubMed
-
- Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Modernising Medical Microbiology Informatics Group, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi:10.1128/AAC.00464-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

