The Impact of Leadered and Leaderless Gene Structures on Translation Efficiency, Transcript Stability, and Predicted Transcription Rates in Mycobacterium smegmatis
- PMID: 32094162
- PMCID: PMC7148126
- DOI: 10.1128/JB.00746-19
The Impact of Leadered and Leaderless Gene Structures on Translation Efficiency, Transcript Stability, and Predicted Transcription Rates in Mycobacterium smegmatis
Abstract
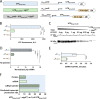
Regulation of gene expression is critical for Mycobacterium tuberculosis to tolerate stressors encountered during infection and for nonpathogenic mycobacteria such as Mycobacterium smegmatis to survive environmental stressors. Unlike better-studied models, mycobacteria express ∼14% of their genes as leaderless transcripts. However, the impacts of leaderless transcript structures on mRNA half-life and translation efficiency in mycobacteria have not been directly tested. For leadered transcripts, the contributions of 5' untranslated regions (UTRs) to mRNA half-life and translation efficiency are similarly unknown. In M. tuberculosis and M. smegmatis, the essential sigma factor, SigA, is encoded by a transcript with a relatively short half-life. We hypothesized that the long 5' UTR of sigA causes this instability. To test this, we constructed fluorescence reporters and measured protein abundance, mRNA abundance, and mRNA half-life and calculated relative transcript production rates. The sigA 5' UTR conferred an increased transcript production rate, shorter mRNA half-life, and decreased apparent translation rate compared to a synthetic 5' UTR commonly used in mycobacterial expression plasmids. Leaderless transcripts appeared to be translated with similar efficiency as those with the sigA 5' UTR but had lower predicted transcript production rates. A global comparison of M. tuberculosis mRNA and protein abundances failed to reveal systematic differences in protein/mRNA ratios for leadered and leaderless transcripts, suggesting that variability in translation efficiency is largely driven by factors other than leader status. Our data are also discussed in light of an alternative model that leads to different conclusions and suggests leaderless transcripts may indeed be translated less efficiently.IMPORTANCE Tuberculosis, caused by Mycobacterium tuberculosis, is a major public health problem killing 1.5 million people globally each year. During infection, M. tuberculosis must alter its gene expression patterns to adapt to the stress conditions it encounters. Understanding how M. tuberculosis regulates gene expression may provide clues for ways to interfere with the bacterium's survival. Gene expression encompasses transcription, mRNA degradation, and translation. Here, we used Mycobacterium smegmatis as a model organism to study how 5' untranslated regions affect these three facets of gene expression in multiple ways. We furthermore provide insight into the expression of leaderless mRNAs, which lack 5' untranslated regions and are unusually prevalent in mycobacteria.
Keywords: 5′ UTR; Mycobacterium tuberculosis; leaderless translation; mRNA stability; posttranscriptional control mechanisms; sigma factors; smegmatis; transcription.
Copyright © 2020 Nguyen et al.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

