HIV-1 uncoats in the nucleus near sites of integration
- PMID: 32094182
- PMCID: PMC7071919
- DOI: 10.1073/pnas.1920631117
HIV-1 uncoats in the nucleus near sites of integration
Abstract
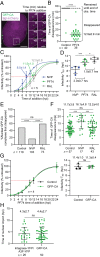
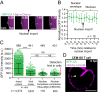
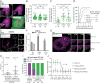
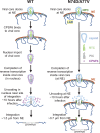
HIV-1 capsid core disassembly (uncoating) must occur before integration of viral genomic DNA into the host chromosomes, yet remarkably, the timing and cellular location of uncoating is unknown. Previous studies have proposed that intact viral cores are too large to fit through nuclear pores and uncoating occurs in the cytoplasm in coordination with reverse transcription or at the nuclear envelope during nuclear import. The capsid protein (CA) content of the infectious viral cores is not well defined because methods for directly labeling and quantifying the CA in viral cores have been unavailable. In addition, it has been difficult to identify the infectious virions because only one of ∼50 virions in infected cells leads to productive infection. Here, we developed methods to analyze HIV-1 uncoating by direct labeling of CA with GFP and to identify infectious virions by tracking viral cores in living infected cells through viral DNA integration and proviral DNA transcription. Astonishingly, our results show that intact (or nearly intact) viral cores enter the nucleus through a mechanism involving interactions with host protein cleavage and polyadenylation specificity factor 6 (CPSF6), complete reverse transcription in the nucleus before uncoating, and uncoat <1.5 h before integration near (<1.5 μm) their genomic integration sites. These results fundamentally change our current understanding of HIV-1 postentry replication events including mechanisms of nuclear import, uncoating, reverse transcription, integration, and evasion of innate immunity.
Keywords: HIV-1; capsid; integration; transcription; uncoating.
Copyright © 2020 the Author(s). Published by PNAS.
Figures

References
-
- Mattei S., Glass B., Hagen W. J., Kräusslich H. G., Briggs J. A., The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

