High HIV-1 diversity in immigrants resident in Italy (2008-2017)
- PMID: 32094387
- PMCID: PMC7039940
- DOI: 10.1038/s41598-020-59084-2
High HIV-1 diversity in immigrants resident in Italy (2008-2017)
Abstract
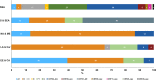
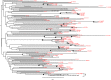
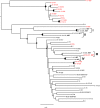
The proportion of new diagnoses of HIV infection in immigrants residing in Italy raised from 11% in 1992 to 29.7% in 2018. To investigate the HIV clades circulating in this community a retrospective study was performed in 557 HIV-infected immigrants living in 12 Italian cities. Immigrants originated from East-Europe and Central-Asia (11.7%), North Africa and Middle East (7.3%), South and South-East Asia (7.2%), Latin America and the Caribbean (14.4%), and sub-Saharan Africa (59.4%). More than 87% of immigrants were on antiretroviral therapy (ART), although 26.6% of them were viremic. A 22.0% of immigrants had hepatitis (HBV and/or HCV) and/or tuberculosis. HIV phylogenetic analysis on sequences from 192 immigrants showed the presence of clades B (23.4%), G (16.1%), C (10.4%), A1 (9.4%), F1 (5.2%), D (1.6%) and Circulating Recombinant Forms (CRFs) (33.9%). CRF02_AG represented 72.3% of the total CRFs. Clusters between immigrants and Italian natives were also present. Drug resistance mutations to NRTI, NNRTI, and PI drug classes occurred in 29.1% of ART-treated and in 12.9% of ART-naïve individuals. These data highlight the need for tailored public health interventions in immigrants to avoid spreading in Italy of HIV genetic forms and ART-resistant variants, as well as HIV co-morbidities.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
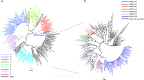


References
-
- ECDC (European Centre for Disease Prevention and Control). Thematic report: Migrants. Monitoring implementation of the Dublin Declaration on Partnership to Fight HIV/AIDS in Europe and Central Asia: 2014 progress report. Stockholm: ECDC, 10.2900/628779 (2015).
-
- ECDC (European Centre for Disease Prevention and Control). HIV and migrants. Monitoring implementation of the Dublin Declaration on Partnership to Fight HIV/AIDS in Europe and Central Asia: 2017 progress report Stockholm: ECDC., 10.2900/063985 (2017).
-
- News Bulletin of Istituto Superiore di Sanità (ISS), Oct, Suppl.1,Vol. 32, N°10, http://old.iss.it/binary/publ/cont/AGGIORNAMENTO_HIV_2018.pdf (2019).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

