A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment
- PMID: 32094587
- PMCID: PMC7104380
- DOI: 10.1038/s41564-020-0674-4
A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment
Abstract
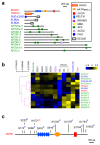
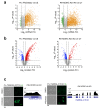
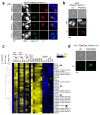
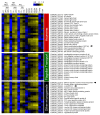
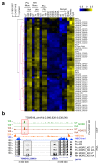
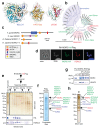
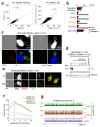
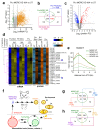
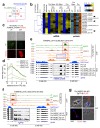
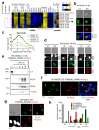
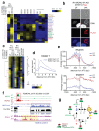
Toxoplasma gondii has a complex life cycle that is typified by asexual development that takes place in vertebrates, and sexual reproduction, which occurs exclusively in felids and is therefore less studied. The developmental transitions rely on changes in the patterns of gene expression, and recent studies have assigned roles for chromatin shapers, including histone modifications, in establishing specific epigenetic programs for each given stage. Here, we identified the T. gondii microrchidia (MORC) protein as an upstream transcriptional repressor of sexual commitment. MORC, in a complex with Apetala 2 (AP2) transcription factors, was shown to recruit the histone deacetylase HDAC3, thereby impeding the accessibility of chromatin at the genes that are exclusively expressed during sexual stages. We found that MORC-depleted cells underwent marked transcriptional changes, resulting in the expression of a specific repertoire of genes, and revealing a shift from asexual proliferation to sexual differentiation. MORC acts as a master regulator that directs the hierarchical expression of secondary AP2 transcription factors, and these transcription factors potentially contribute to the unidirectionality of the life cycle. Thus, MORC plays a cardinal role in the T. gondii life cycle, and its conditional depletion offers a method to study the sexual development of the parasite in vitro, and is proposed as an alternative to the requirement of T. gondii infections in cats.
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Mastering Toxoplasma sex and sleep.Nat Microbiol. 2020 Apr;5(4):533-534. doi: 10.1038/s41564-020-0696-y. Nat Microbiol. 2020. PMID: 32218508 No abstract available.
References
-
- Kim K. The Epigenome, Cell Cycle, and Development in Toxoplasma. 2018:21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

