TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma
- PMID: 32094924
- PMCID: PMC8480332
- DOI: 10.1038/s41591-020-0757-z
TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma
Abstract
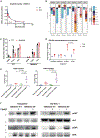
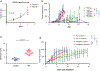
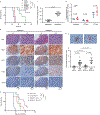
Transmembrane protein 30A (TMEM30A) maintains the asymmetric distribution of phosphatidylserine, an integral component of the cell membrane and 'eat-me' signal recognized by macrophages. Integrative genomic and transcriptomic analysis of diffuse large B-cell lymphoma (DLBCL) from the British Columbia population-based registry uncovered recurrent biallelic TMEM30A loss-of-function mutations, which were associated with a favorable outcome and uniquely observed in DLBCL. Using TMEM30A-knockout systems, increased accumulation of chemotherapy drugs was observed in TMEM30A-knockout cell lines and TMEM30A-mutated primary cells, explaining the improved treatment outcome. Furthermore, we found increased tumor-associated macrophages and an enhanced effect of anti-CD47 blockade limiting tumor growth in TMEM30A-knockout models. By contrast, we show that TMEM30A loss-of-function increases B-cell signaling following antigen stimulation-a mechanism conferring selective advantage during B-cell lymphoma development. Our data highlight a multifaceted role for TMEM30A in B-cell lymphomagenesis, and characterize intrinsic and extrinsic vulnerabilities of cancer cells that can be therapeutically exploited.
Figures












References
-
- Coiffier B et al.CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N. Engl. J. Med 346, 235–242 (2002). - PubMed
-
- Sehn LH et al.Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J. Clin. Oncol 23, 5027–5033 (2005). - PubMed
-
- Alizadeh AA et al.Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). - PubMed
-
- Rosenwald A et al.The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med 346, 1937–1947 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

