Flying Around in the Genome: Characterization of LINE-1 in Chiroptera
- PMID: 32095030
- PMCID: PMC7039574
Flying Around in the Genome: Characterization of LINE-1 in Chiroptera
Abstract
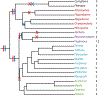
L1s are transposable elements that move by a copy-and-paste mechanism that continuously increases their copy number in the genome, such that each genome has a record of the L1 history in that host lineage. They make up about 20% of the genomes of eutherian mammals and have played a major role in shaping genome evolution. Chiroptera has the lowest average genome size among mammalian orders and the only documented case of L1 extinction affecting an entire mammalian family. Herein, L1 activity and extinction are characterized in all families of the order Chiroptera using a method that enriches for the youngest lineages of L1s in the genome. In addition to the previously reported L1 extinction in Pteropodidae, L1 extinction was documented to occur in Mormoops blainvilli, but this event did not affect all species of Mormoopidae. Further, there was no evidence of concordance between the evolution of L1s and their chiropteran host. There were two L1 lineages present before the divergence of all extant bats. Both lineages are extinct in the Pteropodidae. One or the other L1 lineage is extinct in almost all bat families, but Taphozous melanopogon maintains active members of both. Most intriguingly, some families within the Rhinolophoidea retain one active L1 lineage whereas other families retain the other, creating a deep discontinuity between L1 phylogeny and chiropteran phylogeny. These results indicate that there have been numerous losses of active L1 lineages over the history of chiropteran evolution, but that all chiropteran families except Pteropodidae have retained L1 activity.
Keywords: Chiroptera; L1; LINE-1; bat; evolution; phylogeny; retrotransposons; transposable elements.
Figures


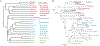
Similar articles
-
Reviving the dead: history and reactivation of an extinct l1.PLoS Genet. 2014 Jun 26;10(6):e1004395. doi: 10.1371/journal.pgen.1004395. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24968166 Free PMC article.
-
Retrofitting the genome: L1 extinction follows endogenous retroviral expansion in a group of muroid rodents.J Virol. 2011 Dec;85(23):12315-23. doi: 10.1128/JVI.05180-11. Epub 2011 Sep 28. J Virol. 2011. PMID: 21957310 Free PMC article.
-
Loss of LINE-1 activity in the megabats.Genetics. 2008 Jan;178(1):393-404. doi: 10.1534/genetics.107.080275. Genetics. 2008. PMID: 18202382 Free PMC article.
-
Chromosomal Evolution in Chiroptera.Genes (Basel). 2017 Oct 13;8(10):272. doi: 10.3390/genes8100272. Genes (Basel). 2017. PMID: 29027987 Free PMC article. Review.
-
L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish.Trends Genet. 2004 Jan;20(1):9-14. doi: 10.1016/j.tig.2003.11.006. Trends Genet. 2004. PMID: 14698614 Review.
Cited by
-
Factors Regulating the Activity of LINE1 Retrotransposons.Genes (Basel). 2021 Sep 30;12(10):1562. doi: 10.3390/genes12101562. Genes (Basel). 2021. PMID: 34680956 Free PMC article. Review.
-
Transposable Elements in Bats Show Differential Accumulation Patterns Determined by Class and Functionality.Life (Basel). 2022 Aug 4;12(8):1190. doi: 10.3390/life12081190. Life (Basel). 2022. PMID: 36013369 Free PMC article.
-
SARS-CoV-2 and human retroelements: a case for molecular mimicry?BMC Genom Data. 2022 Apr 8;23(1):27. doi: 10.1186/s12863-022-01040-2. BMC Genom Data. 2022. PMID: 35395708 Free PMC article.
References
-
- Cantrell MA, Grahn RA, Scott L, and Wichman HA 2000. Isolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques 29:1310–1316. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
