Effectiveness of six molecular typing methods as epidemiological tools for the study of Salmonella isolates in two Colombian regions
- PMID: 32095053
- PMCID: PMC6989315
- DOI: 10.14202/vetworld.2019.1998-2006
Effectiveness of six molecular typing methods as epidemiological tools for the study of Salmonella isolates in two Colombian regions
Abstract
Aim: The aim of this study was the genotypic characterization of the strains of Salmonella spp. isolated from broiler chickens and humans with gastroenteritis from two regions of Colombia, by BOXA1R-polymerase chain reaction (PCR) and random amplification of polymorphic DNA (RAPD)-PCR methods.
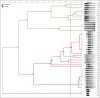
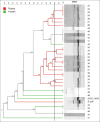
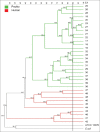
Materials and methods: Forty-nine strains of Salmonella were assessed, 15 from poultry farms in Santander region, and 34 from Tolima region isolated from poultry farms (n=24) and the stool samples of people with gastroenteritis (n=10). BOXA1R primers were selected for repetitive element-based PCR (REP-PCR) and five arbitrary primers, namely, GTG 5, OPB 15, OPP 16, OPS 11, and P 1254 were used for RAPD-PCR to generate DNA fingerprints from the isolates. Fingerprint data from each typing method were under composite analysis and the diversity of the data was analyzed by grouping (clustering). The dendrogram was generated by the unweighted group method with analysis of the arithmetic mean based on the Dice similarity coefficient. In addition, Simpson's index was evaluated to discriminate the power of the methods.
Results: OPP 16 primer and composite analysis proved to be superior compared to other REP-PCR typing methods. The best discriminatory index was observed when GTG 5 (0.92) and OPP 16 (0.85) primers were used alone or combined with RAPD-PCR and BOX-PCR (0.99).
Conclusion: This study indicated that OPP 16 and GTG 5 primers provide suitable molecular typing results for the discrimination of the genetic relationship among Salmonella spp. isolates and may be useful for epidemiological studies.
Keywords: dendrogram; serotyping; typing methods.
Copyright: © Lozano-Villegas, et al.
Figures

References
-
- Eng S.K, Pusparajah P, Ab Mutalib N.S, Ser H.L, Chan K.G, Lee L.H. Salmonella:A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015;8(3):284–293.
-
- Silva C, Betancor L, García C, Astocondor L, Hinostroza N, Bisio J, Rivera J, Perezgasga L, Escanda V, Yim L, Jacobs J, García-del Portillo F, Chabalgoity J, Puente J.L. Characterization of Salmonellaenterica isolates causing bacteremia in Lima, Peru, using multiple typing methods. PloS One. 2017;12(12):e0189946–e0189946. - PMC - PubMed
-
- Wise M.G, Siragusa G.R, Plumblee J, Healy M, Cray P.J, Seal B.S. Predicting Salmonellaenterica serotypes by repetitive sequence-based PCR. J. Microbiol. Methods. 2009;76(1):18–24. - PubMed
-
- Allerberger F, Liesegang A, Grif K, Khaschabi D, Prager R, Danzl J, Höck F, Öttl J, Dierich M.P, Berghold C, Neckstaller I, Tschäpe H, Fisher I. Occurrence of Salmonellaenterica serovar Dublin in Austria. Wien. Med. Wochenschr. 2003;153(7-8):148–152. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
