Comparative modeling and mutual docking of structurally uncharacterized heat shock protein 70 and heat shock factor-1 proteins in water buffalo
- PMID: 32095057
- PMCID: PMC6989329
- DOI: 10.14202/vetworld.2019.2036-2045
Comparative modeling and mutual docking of structurally uncharacterized heat shock protein 70 and heat shock factor-1 proteins in water buffalo
Abstract
Aim: In this study, a wide range of in silico investigation of Bubalus bubalis (BB) heat shock protein 70 (HSP70) and heat shock factor-1 (HSF1) has been performed, ranging from sequence evaluation among species to homology modeling along with their docking studies to decipher the interacting residues of both molecules.
Materials and methods: Protein sequences of BB HSP70 and HSF1 were retrieved from NCBI database in FASTA format. Primary and secondary structure prediction were computed using Expasy ProtParam server and Phyre2 server, respectively. TMHMM server was used to identify the transmembrane regions in HSP70. Multiple sequence alignment and comparative analysis of the protein was carried out using MAFFT and visualization was created using ESPript 3.0. Phylogenetic analysis was accomplished by COBALT. Interactions of HSP70 with other proteins were studied using STRING database. Modeller 9.18, RaptorX, Swiss-Modeller, Phyre2, and I-TASSER were utilized to design the three-dimensional structure of these proteins followed by refinement; energy minimization was accomplished using ModRefiner and SPDBV program. Stereochemical quality along with the accuracy of the predicted models and their visualization was observed by PROCHECK program of PDBsum and UCSF Chimera, respectively. ClusPro 2.0 server was accessed for the docking of the receptor protein with the ligand.
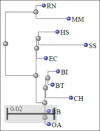
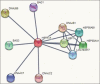
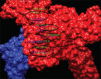
Results: The lower value of Grand Average of Hydropathy indicates the more hydrophilic nature of HSP70 protein. Value of the instability index (II) classified the protein as stable. No transmembrane region was reported for HSP70 by TMHMM server. Phylogenetic analysis based on multiple sequence alignments (MSAs) by COBALT indicated more evolutionarily closeness of Bos indicus (BI) with Bos taurus as compared to BI and BB. STRING database clearly indicates the HSF1 as one of the interacting molecules among 10 interacting partners with HSP 70. The best hit of 3D model of HSP70 protein and HSF1 was retrieved from I-TASSER and Phyre2, respectively. Interacting residues and type of bonding between both the molecules which were docked by ClusPro 2.0 were decoded by PIC server. Hydrophobic interactions, protein-protein main-chain-side-chain hydrogen bonds, and protein-protein side-chain-side-chain hydrogen bonds were delineated in this study.
Conclusion: This is the first-ever study on in silico interaction of HSP70 and HSF1 proteins in BB. Several bioinformatics web tools were utilized to study secondary structure along with comparative modeling, physicochemical properties, and protein-protein interaction. The various interacting amino acid residues of both proteins have been indicated in this study.
Keywords: Bubalus bubalis; docking; heat shock factor-1; heat shock protein 70; heat shock proteins; homology modeling.
Copyright: © Singh, et al.
Figures






References
-
- Creagh E.M, Carmody R.J, Cotter T.G. Heat shock protein 70 inhibits caspase-dependent and-independent apoptosis in Jurkat T cells. Exp. Cell. Res. 2000;257(1):58–66. - PubMed
-
- Lindquist S, Craig E.A. The heat-shock proteins. Annu. Rev. Genet. 1988;22(1):631–677. - PubMed
-
- Archana P.R, Aleena J, Pragna P, Vidya M.K, Niyas A.P.A, Bagath M, Krishnan G, Manimaran A, Beena V, Kurien E.K, Sejian V. Role of heat shock proteins in livestock adaptation to heat stress. J. Dairy Vet. Anim. Res. 2017;5(1):00127.
-
- Pawar H.N, Kumar G.R, Narang R, Agrawal R.K. Heat and cold stress enhances the expression of heat shock protein 70, heat shock transcription factor 1 and cytokines (IL-12, TNF-and GMCSF) in buffaloes. Int. J. Curr. Microbiol. Appl. Sci. 2014;3(2):307–317.
LinkOut - more resources
Full Text Sources
