Human RAP1 specifically protects telomeres of senescent cells from DNA damage
- PMID: 32096305
- PMCID: PMC7132343
- DOI: 10.15252/embr.201949076
Human RAP1 specifically protects telomeres of senescent cells from DNA damage
Abstract
Repressor/activator protein 1 (RAP1) is a highly evolutionarily conserved protein found at telomeres. Although yeast Rap1 is a key telomere capping protein preventing non-homologous end joining (NHEJ) and consequently telomere fusions, its role at mammalian telomeres in vivo is still controversial. Here, we demonstrate that RAP1 is required to protect telomeres in replicative senescent human cells. Downregulation of RAP1 in these cells, but not in young or dividing pre-senescent cells, leads to telomere uncapping and fusions. The anti-fusion effect of RAP1 was further explored in a HeLa cell line where RAP1 expression was depleted through an inducible CRISPR/Cas9 strategy. Depletion of RAP1 in these cells gives rise to telomere fusions only when telomerase is inhibited. We further show that the fusions triggered by RAP1 loss are dependent upon DNA ligase IV. We conclude that human RAP1 is specifically involved in protecting critically short telomeres. This has important implications for the functions of telomeres in senescent cells.
Keywords: RAP1; chromosome fusions; non-homologous end joining; replicative senescence; telomeres.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
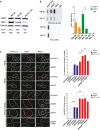
Western blotting showing the expression of RAP1 and TRF2 in MRC‐5 cells of different population doublings (PD). Senescent cells (PD 72) were left in culture for at least 4 weeks before harvesting for analysis.
ChIP analysis of young (PD 30) and senescent (PD 72) MRC‐5 cells performed with either anti‐RAP1, anti‐TRF2, or an IgG antibody. The immunoprecipitated and input products were loaded into a slot blot membrane and hybridized with a telomere probe, and stripped and hybridized to an Alu probe in order to determine unspecific binding. Quantification was performed by normalizing the telomere signal of the immunoprecipitated to that of the input. Data represent mean ± SD of three biological replicates.
Immunofluorescence detection of 53BP1 (red) and a FISH probe staining telomeres (green) in young (PD 26), pre‐senescent (PD 66), and senescent (PD 72 + 4 weeks) MRC‐5 fibroblasts. The upper graph shows the percentage of telomere co‐localizing with 53BP1 (TIFs). Total DNA damage was measured by immunofluorescence detection of 53BP1. The lower graph shows the total number of 53BP1 foci per nucleus. Approximately 40–50 cells were analyzed per replicate and per condition. Bars represent SEM of two biological replicates.
RAP1 expression of dividing MRC‐5 cells (left panel; PD 26 and PD 66) and senescent cells (right panel; PD72 + 4 weeks). RAP1 depletion (or ectopic expression—right panel) was carried out for 10 days.
Telomere restriction fragment of young (PD 26) and senescent cells (PD 72 + 4 weeks) upon 10‐day shRAP1 incubation.
Single telomere length analysis (STELA) of MRC‐5 senescent cells transduced with either a control vector or an shRAP1‐expressing vector. STELA was performed at the Xp/Yp telomere, and the mean telomere length ± SD, after subtracting 406 bp of flanking DNA, is shown at the bottom of the blot.

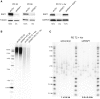
- A
Location of the primers and probes used for the telomere fusion PCR: Red arrows indicate positions of subtelomeric PCR primers, while black arrows represent the primers used to generate DNA probes for radioactive hybridization with the Southern blot membranes.
- B, C
Representative Southern blot membranes of the telomere fusion PCR assay shown in Fig 2A and B. Membranes were subsequently hybridized with the 16p probe, XpYp probe, and finally with the 21q probe.
- D
Different RAP1 constructs used in the telomere fusion assay of senescent MRC‐5 cells. The percentage of protein expression is indicated below the blots.
- E
Expression of RAP1, LIG3, and LIG4 related to Fig 2C. The percentage of protein expression is indicated.

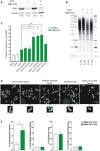
- A
Number of telomere fusions performed by PCR using 3 subtelomeric primers in the same reaction (16p1, 21q1, and XpYpM). The primer 16p1 is able to bind the subtelomeric region of 1p 9q, 12p, 15q, 16p, XqYq, and the interstitial 2q14 region; the 21q1 primer binds the subtelomeric region of 1q, 2q, 5q, 6q, 6p, 8p, 10q, 13q, 19p, 19q, 21q, 22q, and the interstitial 2q13 site. The PCR‐fusion assay was performed in MRC‐5 primary fibroblasts of different population doublings (PD) with or without depletion of RAP1 (shRAP1 or shControl, respectively). Lentiviral infection was performed for 10 days. Data represent mean ± SD of three biological replicates. Statistical analyses were performed using Mann–Whitney U‐test (**P < 0.001; ***P < 0.0001).
- B
Southern blotting showing the hybridization of the 16p probe for the conditions described in (A). Number of fusions per 1 μg of DNA is shown at the bottom of the gels.
- C, D
Number of fusions per 1 μg of DNA in MRC‐5 senescent cells. Lentiviral transfections with shRNAs were carried out for 10 days while transfections with siRNAs for 6 days (two subsequent transfections of 3 days each). Representative Southern blot membranes hybridized with the 16p probe are shown. Data represent mean ± SD of three biological replicates. Statistical analyses were performed using Mann–Whitney U‐test (*P < 0.05; **P < 0.001; ***P < 0.0001).

Distribution of the telomere repeat array found at the junction between two fused chromosomes of senescent cells transduced with lentiviral vectors expressing either shControl or shRAP1. Variant telomere repeats were included in the estimation of the telomere array length. Data represent median ± interquartile range (green lines) of three biological replicates. Statistical analysis was performed using Mann–Whitney U‐test ***P < 0.0001).
Examples of DNA sequences found at the telomere fusion points of shControl and shRAP1 conditions. Telomere repeats are underlined in blue, while variant telomere repeats are underlined in red. The sequence ID corresponds to the identifier in Table EV1.
RAP1 expression in HeLa cells treated with the telomerase inhibitor BIBR1532 for 25 days (20 μM final concentration). Fifteen days before cell harvesting, doxycycline (DOX; 1 μg/μl final concentration) was added to deplete the expression of RAP1.
Telomere length analysis by Southern blotting of the samples described in (A). The size of the main intensity peak is indicated at the bottom of the gel.
Number of fusions in HeLa cells after 25 days in culture. Cells were maintained with BIBR1532 during the whole period of the experiment, while doxycycline (1 μg/μl final concentration) and the infection with shControl, shLIG3, or shLIG4 were carried out for the last 15 days of the experiment. Data represent mean ± SD of three biological replicates. Statistical analyses were performed using Mann–Whitney U‐test (*P < 0.05; **P < 0.001; ***P < 0.0001).
Examples of metaphase spreads hybridized with a telomeric PNA probe (green) when RAP1 was depleted (+DOX). Telomerase was inhibited with BIBR1532 for 25 days. Scale bar = 10 μm.
Quantification of chromosome aberrations observed in RAP1‐depleted cells (+DOX) or control (‐DOX). Data represent mean ± SD of three biological replicates. Approximately 15 metaphase spreads were analyzed per replicate with a total of 2,300 chromosomes examined per condition. *P < 0.05; two‐tailed Student's t‐test.
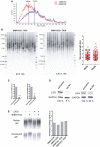
Telomere signal profile of cells treated with or without BIBR1532 +DOX as shown in Fig 4B. The size in kilobases of the highest peak is indicated.
STELA assay at the Xp/Yp telomere of HeLa cells treated with the telomerase inhibitor BIBR1532 and depleted for RAP1 (+DOX). The scatter plots show the distribution of STELA products after subtracting 406 bp of flanking DNA, while error bars represent the mean ± standard deviation.
Relative mRNA levels of LIG3 and LIG4 measured by RT–qPCR corresponding to the experiment shown in Fig 4C. Data represent mean ± SD of three independent experiments (***P = 0.0001 for shLIG3 and **P = 0.0061 for shLIG4; two‐tailed Student's t‐test).
Protein expression of LIG3 and LIG4 used in Fig 4C. The percentage of protein expression is indicated.
Telomere overhang assay in HeLa cells. In‐gel hybridization of the telomere probe in native and denaturing conditions. On the right, quantification of the normalized telomere overhang signal performed by dividing the signal intensity obtained by native gel hybridization to the total signal obtained with denatured gels. Data represent one biological replicate.

Growth curve of MRC‐5 fibroblasts. Cells were grown until senescence and after 3 weeks of no growth detection, lentiviral infections containing either shp21CIP1 or shp21CIP1 + shRAP1 sequences were performed. Cells were harvested 15 days post‐infection (dpi). Data represent mean ± SD of three biological replicates measured after 8 and 15 dpi.
Expression of p21CIP1 and RAP1 after 15 dpi of cells transduced with shControl, shp21CIP1, or shp21CIP1 + shRAP1.
Dividing cells (magenta) were identified by EdU staining (1 μM EdU for 24 h). The percentage of dividing cells after 15 dpi is shown (n = 3). Scale bar = 10 μm.
Senescence‐associated β‐galactosidase (SA‐β‐gal) staining in senescent and post‐senescent fibroblasts (1 and 15 dpi) is shown. Percentage of SA‐β‐gal‐positive cells is indicated for each condition. Three biological replicates were performed, with approximately 300 cells analyzed per condition. Scale bar = 50 μm.
Senescent cells, transduced with shp21CIP1 or shp21CIP1 + shRAP1 for 15 days, were stained with CytoCalcein violet (live cells), Apopxin Green (apoptotic cells), and 7‐aminoactinomycin D (7‐AAD) (late apoptotic/necrotic cells) and visualized by flow cytometry.
Quantification of apoptotic cells from the conditions described in (E). Data represent mean ± SD of three biological replicates (**P < 0.001; two‐tailed Student's t‐test).
References
-
- Li B, Oestreich S, de Lange T (2000) Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- 81522017/National Natural Science Foundation of China/International
- ANR-19-CE12-0020-01/Agence Nationale de la Recherche (ANR) (program TELOCHROM)/International
- 19XD1422500/Shanghai Municipal Science and Technology Commission/International
- G0500336/MRC_/Medical Research Council/United Kingdom
- Institut Nationale du Cancer (INCa) (program REPLITOP)/International
- 81971312/National Natural Science Foundation of China/International
- FDT20170437327/Fondation pour la Recherche Médicale FRM/International
- 91749126/National Natural Science Foundation of China/International
- 15ZH4005/Foundation of Shanghai Jiaotong University School of Medicine for Translational Medicine Innovation Project/International
- ANR-11-LABX-0028-01/LABEX SIGNALIFE/International
- Fondation ARC ("Programme Labellisé")/International
LinkOut - more resources
Full Text Sources
Research Materials

