Understanding the human antibody repertoire
- PMID: 32097086
- PMCID: PMC7153836
- DOI: 10.1080/19420862.2020.1729683
Understanding the human antibody repertoire
Abstract
The origins of the various elements in the human antibody repertoire have been and still are subject to considerable uncertainty. Uncertainty in respect of whether the various elements have always served a specific defense function or whether they were co-opted from other organismal roles to form a crude naïve repertoire that then became more complex as combinatorial mechanisms were added. Estimates of the current size of the human antibody naïve repertoire are also widely debated with numbers anywhere from 10 million members, based on experimentally derived numbers, to in excess of one thousand trillion members or more, based on the different sequences derived from theoretical combinatorial calculations. There are questions that are relevant at both ends of this number spectrum. At the lower bound it could be questioned whether this is an insufficient repertoire size to counter all the potential antigen-bearing pathogens. At the upper bound the question is rather simpler: How can any individual interrogate such an astronomical number of antibody-bearing B cells in a timeframe that is meaningful? This review evaluates the evolutionary aspects of the adaptive immune system, the calculations that lead to the large repertoire estimates, some of the experimental evidence pointing to a more restricted repertoire whose variation appears to derive from convergent 'structure and specificity features', and includes a theoretical model that seems to support it. Finally, a solution that may reconcile the size difference anomaly, which is still a hot subject of debate, is suggested.
Keywords: Antibody repertoire; adaptive immunity; infinite repertoire theorem; naïve repertoire; variable region assembly.
Figures


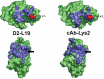
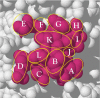
References
-
- Flajnik FM, Du Pasquier L.. Evolution of the immune system. In: Paul WE, editor. Fundamental immunology. 7th ed. Lippincot (PA): Williams & Wilkins; 2013. p. 67–16.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
