Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART
- PMID: 32097124
- PMCID: PMC7213790
- DOI: 10.1172/jci.insight.134196
Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART
Abstract
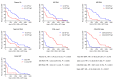
Plasma viral load (VL) and CD4+ T cell count are widely used as biomarkers of HIV type 1 (HIV-1) replication, pathogenesis, and response to antiretroviral therapy (ART). However, the clinical potential of cell-associated (CA) HIV-1 molecular markers is much less understood. Here, we measured CA HIV-1 RNA and DNA in HIV-infected individuals treated with temporary ART initiated during primary HIV-1 infection. We demonstrate substantial predictive value of CA RNA for (a) the virological and immunological response to early ART, (b) the magnitude and time to viral rebound after discontinuation of early ART, and (c) disease progression in the absence of treatment. Remarkably, when adjusted for CA RNA, plasma VL no longer appeared as an independent predictor of any clinical endpoint in this cohort. The potential of CA RNA as an HIV-1 clinical marker, in particular as a predictive biomarker of virological control after stopping ART, should be explored in the context of HIV-1 curative interventions.
Keywords: AIDS/HIV; Transcription; Virology.
Conflict of interest statement
Figures




References
-
- Pasternak AO, Jurriaans S, Bakker M, Prins JM, Berkhout B, Lukashov VV. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12):e8490. doi: 10.1371/journal.pone.0008490. - DOI - PMC - PubMed
-
- European Pregnancy Paediatric HIV Cohort Collaboration (EPPICC) Early-treated Perinatally HIV-infected Individuals: Improving Children’s Actual Life with Novel Immunotherapeutic Strategies (EPIICAL) study groups. Predictors of faster virological suppression in early treated infants with perinatal HIV from Europe and Thailand. AIDS. 2019;33(7):1155–1165. doi: 10.1097/QAD.0000000000002172. - DOI - PMC - PubMed
-
- Ceccherini-Silberstein F, et al. Pre-ART HIV-1 DNA in CD4+ T cells correlates with baseline viro-immunological status and outcome in patients under first-line ART. J Antimicrob Chemother. 2018;73(12):3460–3470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

