Regulators of an ancient polyphenism evolved through episodic protein divergence and parallel gene radiations
- PMID: 32098612
- PMCID: PMC7062019
- DOI: 10.1098/rspb.2019.2595
Regulators of an ancient polyphenism evolved through episodic protein divergence and parallel gene radiations
Abstract
Polyphenism is a form of developmental plasticity that transduces environmental cues into discontinuous, often disparate phenotypes. In some cases, polyphenism has been attributed to facilitating morphological diversification and even the evolution of novel traits. However, this process is predicated on the origins and evolutionary maintenance of genetic mechanisms that specify alternate developmental networks. When and how regulatory loci arise and change, specifically before and throughout the history of a polyphenism, is little understood. Here, we establish a phylogenetic and comparative molecular context for two dynamically evolving genes, eud-1 and seud-1, which regulate polyphenism in the nematode Pristionchus pacificus. This species is dimorphic in its adult feeding-structures, allowing individuals to become microbivores or facultative predators depending on the environment. Although polyphenism regulation is increasingly well understood in P. pacificus, the polyphenism is far older than this species and has diversified morphologically to enable an array of ecological functions across polyphenic lineages. To bring this taxonomic diversity into a comparative context, we reconstructed the histories of eud-1 and seud-1 relative to the origin and diversification of polyphenism, finding that homologues of both genes have undergone lineage-specific radiations across polyphenic taxa. Further, we detected signatures of episodic diversifying selection on eud-1, particularly in early diplogastrid lineages. Lastly, transgenic rescue experiments suggest that the gene's product has functionally diverged from its orthologue's in a non-polyphenic outgroup. In summary, we provide a comparative framework for the molecular components of a plasticity switch, enabling studies of how polyphenism, its regulation, and ultimately its targets evolve.
Keywords: developmental switch; gene duplication; nematodes; phenotypic plasticity; sulfotransferase; sulphatase.
Conflict of interest statement
The authors declare no competing interests.
Figures
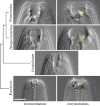
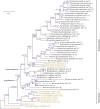

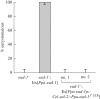
References
-
- West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
