Dual-molecular barcode sequencing detects rare variants in tumor and cell free DNA in plasma
- PMID: 32099048
- PMCID: PMC7042261
- DOI: 10.1038/s41598-020-60361-3
Dual-molecular barcode sequencing detects rare variants in tumor and cell free DNA in plasma
Abstract
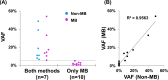
Conventional next generation sequencing analysis has provided important insights into cancer genetics. However, the detection of rare (low allele fraction) variants remains difficult because of the error-prone nucleotide changes derived from sequencing/PCR errors. To eliminate the false-positive variants and detect genuine rare variants, sequencing technology combined with molecular barcodes will be useful. Here, we used the newly developed dual-molecular barcode technology (Ion AmpliSeq HD) to analyze somatic mutations in 24 samples (12 tumor tissues and 12 plasma) from 12 patients with biliary-pancreatic and non-small cell lung cancers. We compared the results between next generation sequencing analysis with or without molecular barcode technologies. The variant allele fraction (VAF) between non-molecular barcode and molecular barcode sequencing was correlated in plasma DNA (R2 = 0.956) and tumor (R2 = 0.935). Both methods successfully detected high VAF mutations, however, rare variants were only identified by molecular barcode sequencing and not by non-molecular barcode sequencing. Some of these rare variants in tumors were annotated as pathogenic, and therefore subclonal driver mutations could be observed. Furthermore, the very low VAF down to 0.17% were identified in cell free DNA in plasma. These results demonstrate that the dual molecular barcode sequencing technologies can sensitively detect rare somatic mutations, and will be important in the investigation of the clonal and subclonal architectures of tumor heterogeneity.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

