Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa
- PMID: 32099423
- PMCID: PMC7006860
- DOI: 10.2147/IDR.S238811
Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa
Abstract
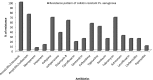
Background and aim: The emergence of colistin-resistant strains is considered a great threat for patients with severe infections. Here, we investigate the prevalence and some possible mechanisms of colistin resistance among multidrug-resistant (MDR) and extensively drug-resistant (XDR) Pseudomonas aeruginosa (P. aeruginosa).
Methods: Antimicrobial susceptibility was performed using disc diffusion methods while colistin resistance was detected by agar dilution method. Possible mechanisms for colistin resistance were studied by detection of mcr-1 and mcr-2 genes by conventional PCR, detection of efflux mechanisms using Carbonyl Cyanide 3-Chlorophenylhydrazone (CCCP), studying outer membrane protein profile and Lipopolysaccharide (LPS) profile of resistant isolates.
Results: It was found that MDR and XDR represented 96% and 87% of the isolated P. aeruginosa, respectively, and colistin resistance represented 21.3%. No isolates were positive for mcr-2 gene while 50% of colistin-resistant isolates were positive for mcr-1. Efflux mechanisms were detected in 3 isolates. Protein profile showed the presence of a band of 21.4 KDa in the resistant strains which may represent OprH while LPS profile showed differences among colistin-resistant mcr-1 negative strains, colistin-resistant mcr-1 positive strains and susceptible strains.
Conclusion: The current study reports a high prevalence of colistin resistance and mcr-1 gene in P. aeruginosa strains isolated from Egypt that may result in untreatable infections. Our finding makes it urgent to avoid unnecessary clinical use of colistin.
Keywords: MDR; Pseudomonas aeruginosa; XDR; colistin resistance; mcr-1; mcr-2; toxA gene.
© 2020 Abd El-Baky et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures




References
-
- Farhan SM, Ibrahim RA, Mahran KM, Hetta HF, El-Baky RMA. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect Drug Resist. 2019;12:2125. doi: 10.2147/IDR.S198373 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

