Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity
- PMID: 32101166
- PMCID: PMC7159879
- DOI: 10.7554/eLife.52696
Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity
Abstract
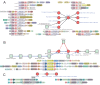
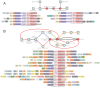
Social cellular aggregation or multicellular organization pose increased risk of transmission of infections through the system upon infection of a single cell. The generality of the evolutionary responses to this outside of Metazoa remains unclear. We report the discovery of several thematically unified, remarkable biological conflict systems preponderantly present in multicellular prokaryotes. These combine thresholding mechanisms utilizing NTPase chaperones (the MoxR-vWA couple), GTPases and proteolytic cascades with hypervariable effectors, which vary either by using a reverse transcriptase-dependent diversity-generating system or through a system of acquisition of diverse protein modules, typically in inactive form, from various cellular subsystems. Conciliant lines of evidence indicate their deployment against invasive entities, like viruses, to limit their spread in multicellular/social contexts via physical containment, dominant-negative interactions or apoptosis. These findings argue for both a similar operational 'grammar' and shared protein domains in the sensing and limiting of infections during the multiple emergences of multicellularity.
Keywords: AAA+ ATPase; DEATH domain; GTPase; apoptosis; chaperones; genetics; genomics; infectious disease; microbiology; multicellular prokaryotes.
Plain language summary
Bacteria are the most numerous lifeforms on the planet. Most bacteria live as single cells that grow and multiply independently within larger communities of microbes. However, some bacterial cells assemble to form more complex structures where individual cells might perform distinct roles. Such bacteria are referred to as ‘multicellular bacteria’. For example, cells of bacteria known as Streptomyces collectively form filaments that help the bacteria collect nutrients from their food sources, and aerial structures bearing reproductive spores. Bacteria in these filaments may come into contact with many other microbes in their surroundings including other bacteria within the same filament, other species of bacteria, and viruses. These contacts often lead to conflict, for example, if the microbes compete with each other for nutrients or if a virus tries to attack the bacteria. Bacteria have evolved immune systems that detect other microbes and use antibiotics, toxins and other defense mechanisms against them. Compared to single-celled bacteria, multicellular bacteria may be more vulnerable to threats from viruses because once a virus has overcome the defenses of one cell in the multicellular assembly, it may be easier for it to kill, or spread to the other cells. However, it is not clear how these systems evolved to deal with the unique problems of multicellular bacteria. Now, Kaur, Burroughs et al. have used computational approaches to search for new immune systems in diverse multicellular bacteria. The new classes of systems they found are each made of different molecular components, but all require a large input of energy to be activated. This activation barrier prevents the bacterial cells from deploying weapons unless the signal from the enemy microbe crosses a high enough threshold. Many tools used in molecular biology, and increasingly in medicine, have been derived from the immune systems of bacteria, such as the enzymes that cut or edit DNA. The findings of Kaur, Burroughs et al. may aid the development of new tools that specifically bind to viruses or other dangerous microbes, or inhibit their ability to interact with components in cells. The next step would be to perform experiments using some of the immune systems identified in this work.
Conflict of interest statement
GK, AB, LI, LA No competing interests declared
Figures





References
-
- Alayyoubi M, Guo H, Dey S, Golnazarian T, Brooks GA, Rong A, Miller JF, Ghosh P. Structure of the essential diversity-generating retroelement protein bAvd and its functionally important interaction with reverse transcriptase. Structure. 2013;21:266–276. doi: 10.1016/j.str.2012.11.016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

