Human RPA activates BLM's bidirectional DNA unwinding from a nick
- PMID: 32101168
- PMCID: PMC7065910
- DOI: 10.7554/eLife.54098
Human RPA activates BLM's bidirectional DNA unwinding from a nick
Abstract
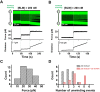
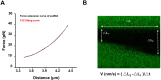
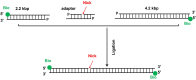
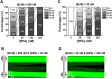
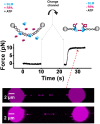
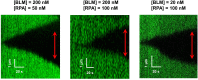
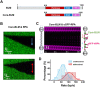
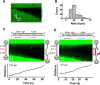
BLM is a multifunctional helicase that plays critical roles in maintaining genome stability. It processes distinct DNA substrates, but not nicked DNA, during many steps in DNA replication and repair. However, how BLM prepares itself for diverse functions remains elusive. Here, using a combined single-molecule approach, we find that a high abundance of BLMs can indeed unidirectionally unwind dsDNA from a nick when an external destabilizing force is applied. Strikingly, human replication protein A (hRPA) not only ensures that limited quantities of BLMs processively unwind nicked dsDNA under a reduced force but also permits the translocation of BLMs on both intact and nicked ssDNAs, resulting in a bidirectional unwinding mode. This activation necessitates BLM targeting on the nick and the presence of free hRPAs in solution whereas direct interactions between them are dispensable. Our findings present novel DNA unwinding activities of BLM that potentially facilitate its function switching in DNA repair.
Keywords: BLM; DNA unwinding; RPA; chicken; helicase; molecular biophysics; optical tweezers; single molecule; structural biology.
© 2020, Qin et al.
Conflict of interest statement
ZQ, LB, XH, SZ, XZ, YL, ML, MM, XX, BS No competing interests declared
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
- 2016YFA0500902/Ministry of Science and Technology of the People's Republic of China
- 2017YFA0106700/Ministry of Science and Technology of the People's Republic of China
- 19ZR1434100/Shanghai Natural Science Foundation
- PLBIO2017-167/Institut National Du Cancer
- EL2028.LNCC/MaM/The French National League Against Cancer
LinkOut - more resources
Full Text Sources

