Discovering and interpreting transcriptomic drivers of imaging traits using neural networks
- PMID: 32101278
- PMCID: PMC7267841
- DOI: 10.1093/bioinformatics/btaa126
Discovering and interpreting transcriptomic drivers of imaging traits using neural networks
Abstract
Motivation: Cancer heterogeneity is observed at multiple biological levels. To improve our understanding of these differences and their relevance in medicine, approaches to link organ- and tissue-level information from diagnostic images and cellular-level information from genomics are needed. However, these 'radiogenomic' studies often use linear or shallow models, depend on feature selection, or consider one gene at a time to map images to genes. Moreover, no study has systematically attempted to understand the molecular basis of imaging traits based on the interpretation of what the neural network has learned. These studies are thus limited in their ability to understand the transcriptomic drivers of imaging traits, which could provide additional context for determining clinical outcomes.
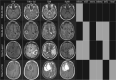
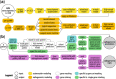
Results: We present a neural network-based approach that takes high-dimensional gene expression data as input and performs non-linear mapping to an imaging trait. To interpret the models, we propose gene masking and gene saliency to extract learned relationships from radiogenomic neural networks. In glioblastoma patients, our models outperformed comparable classifiers (>0.10 AUC) and our interpretation methods were validated using a similar model to identify known relationships between genes and molecular subtypes. We found that tumor imaging traits had specific transcription patterns, e.g. edema and genes related to cellular invasion, and 10 radiogenomic traits were significantly predictive of survival. We demonstrate that neural networks can model transcriptomic heterogeneity to reflect differences in imaging and can be used to derive radiogenomic traits with clinical value.
Availability and implementation: https://github.com/novasmedley/deepRadiogenomics.
Contact: whsu@mednet.ucla.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures







References
-
- Bengio Y. (2009) Learning Deep Architectures for AI, Vol. 2. Now Publishers, Inc, Boston, MA, USA.

