A convenient renewable surface plasmon resonance chip for relative quantification of genetically modified soybean in food and feed
- PMID: 32101588
- PMCID: PMC7043770
- DOI: 10.1371/journal.pone.0229659
A convenient renewable surface plasmon resonance chip for relative quantification of genetically modified soybean in food and feed
Abstract
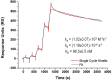
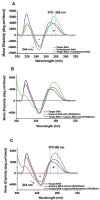
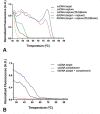
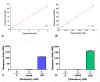
The cultivation of genetically modified organisms (GMO) continues to expand worldwide. Still, many consumers express concerns about the use of GMO in food or feed, and many countries have legislated on labelling systems to indicate the presence of GMO in commercial products. To deal with the increased number of GMO events and to address related regulations, alternative detection methods for GMO inspection are required. In this work, a genosensor based on Surface Plasmon Resonance under continuous flow was developed for the detection and quantification of a genetically modified soybean (event GTS 40-3-2). In a single chip, the simultaneous detection of the event-specific and the taxon-specific samples were achieved, whose detection limits were 20 pM and 16 pM, respectively. The reproducibility was 1.4%, which supports the use of the chip as a reliable and cost-effective alternative to other DNA-based techniques. The results indicate that the proposed method is a versatile tool for GMO quantification in food and feed samples.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Use of pJANUS-02-001 as a calibrator plasmid for Roundup Ready soybean event GTS-40-3-2 detection: an interlaboratory trial assessment.Anal Bioanal Chem. 2010 Mar;396(6):2165-73. doi: 10.1007/s00216-009-3346-z. Epub 2009 Dec 17. Anal Bioanal Chem. 2010. PMID: 20016879 Free PMC article.
-
Current and new approaches in GMO detection: challenges and solutions.Biomed Res Int. 2015;2015:392872. doi: 10.1155/2015/392872. Epub 2015 Oct 15. Biomed Res Int. 2015. PMID: 26550567 Free PMC article. Review.
-
Detection of Soybean GMO Events Using Two Multiplex Droplet Digital PCR Assays.J AOAC Int. 2025 Jan 1;108(1):23-28. doi: 10.1093/jaoacint/qsae082. J AOAC Int. 2025. PMID: 39475430 Free PMC article.
-
[Detection of genetically modified organisms in food and animal feed by polymerase chain reaction].Wei Sheng Yan Jiu. 2005 Nov;34(6):732-4. Wei Sheng Yan Jiu. 2005. PMID: 16535848 Chinese.
-
Detection and traceability of genetically modified organisms in the food production chain.Food Chem Toxicol. 2004 Jul;42(7):1157-80. doi: 10.1016/j.fct.2004.02.018. Food Chem Toxicol. 2004. PMID: 15123385 Review.
References
-
- Gupta R, Singh RL. Genetically modified organisms (GMOs) and environment. Principles and Applications of Environmental Biotechnology for a Sustainable Future. Singapore: Springer; 2017. p. 425–465. 10.1007/978-981-10-1866-4. - DOI
-
- International Service for the Acquisition of Agri-biotech Applications (ISAAA). Global Status of Commercialized Biotech/GM Crops: 2016. ISAAA Brief No. 52. New York: ISAAA, 2016.
-
- Windels P, Taverniers I, Depicker A, Van Bockstaele E, De Loose M. Characterisation of the Roundup Ready soybean insert. Eur Food Res Technol. 2001; 213:107–112. 10.1007/s002170100336. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

