Probing RNA Conformational Equilibria within the Functional Cellular Context
- PMID: 32101729
- PMCID: PMC7941409
- DOI: 10.1016/j.celrep.2020.02.004
Probing RNA Conformational Equilibria within the Functional Cellular Context
Abstract
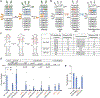
Low-abundance short-lived non-native conformations referred to as excited states (ESs) are increasingly observed in vitro and implicated in the folding and biological activities of regulatory RNAs. We developed an approach for assessing the relative abundance of RNA ESs within the functional cellular context. Nuclear magnetic resonance (NMR) spectroscopy was used to estimate the degree to which substitution mutations bias conformational equilibria toward the inactive ES in vitro. The cellular activity of the ES-stabilizing mutants was used as an indirect measure of the conformational equilibria within the functional cellular context. Compensatory mutations that restore the ground-state conformation were used to control for changes in sequence. Using this approach, we show that the ESs of two regulatory RNAs from HIV-1, the transactivation response element (TAR) and the Rev response element (RRE), likely form in cells with abundances comparable to those measured in vitro, and their targeted stabilization may provide an avenue for developing anti-HIV therapeutics.
Keywords: HIV-1 transactivation response element; RNA drug discovery; RNA dynamics; RNA structure; RRE; Rev response element; TAR; cellular activity; conformational switches; excited state; structure mapping; transactivation.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests H.M.A.-H. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company. He is co-author on the patent “High-Throughput Ensemble-Based Docking Against Flexible Biomolecular Targets” (8,498,823).
Figures


References
-
- Bartel DP, Zapp ML, Green MR, Szostak JW (1991). HIV-1 Rev Regulation Involves Recognition of Non-Watson-Crick Base Pairs in Viral RNA. Cell 67, 529–536. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

