DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs
- PMID: 32102413
- PMCID: PMC7070539
- DOI: 10.3390/molecules25041015
DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs
Abstract
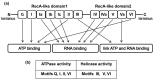
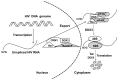
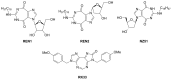
This short review is focused on enzymatic properties of human ATP-dependent RNA helicase DDX3 and the development of antiviral and anticancer drugs targeting cellular helicases. DDX3 belongs to the DEAD-box proteins, a large family of RNA helicases that participate in all aspects of cellular processes, such as cell cycle progression, apoptosis, innate immune response, viral replication, and tumorigenesis. DDX3 has a variety of functions in the life cycle of different viruses. DDX3 helicase is required to facilitate both the Rev-mediated export of unspliced/partially spliced human immunodeficiency virus (HIV) RNA from nucleus and Tat-dependent translation of viral genes. DDX3 silencing blocks the replication of HIV, HCV, and some other viruses. On the other hand, DDX displays antiviral effect against Dengue virus and hepatitis B virus through the stimulation of interferon beta production. The role of DDX3 in different types of cancer is rather controversial. DDX3 acts as an oncogene in one type of cancer, but demonstrates tumor suppressor properties in other types. The human DDX3 helicase is now considered as a new attractive target for the development of novel pharmaceutical drugs. The most interesting inhibitors of DDX3 helicase and the mechanisms of their actions as antiviral or anticancer drugs are discussed in this short review.
Keywords: DEAD-box family RNA helicases; anticancer drug; antiviral drug; inhibitors; physico-chemical properties; virus life cycle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
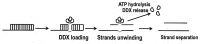
References
-
- Xi X.G. Helicases as antiviral and anticancer drug targets. Curr. Med. Chem. 2007;14:883–915. - PubMed
-
- Maga G., Falchi F., Radi M., Botta L., Casaluce G., Bernardini M., Irannejad H., Manetti F., Garbelli A., Samuele A., et al. Toward the discovery of novel anti-HIV drugs. Second-generation inhibitors of the cellular ATPase DDX3 with improved anti-HIV activity: Synthesis, structure-activity relationship analysis, cytotoxicity studies, and target validation. ChemMedChem. 2011;6:1371–1389. doi: 10.1002/cmdc.201100166. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

