Nrp1 is Activated by Konjac Ceramide Binding-Induced Structural Rigidification of the a1a2 Domain
- PMID: 32102436
- PMCID: PMC7072815
- DOI: 10.3390/cells9020517
Nrp1 is Activated by Konjac Ceramide Binding-Induced Structural Rigidification of the a1a2 Domain
Abstract
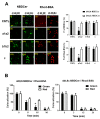
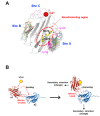
Konjac ceramide (kCer) is a plant-type ceramide composed of various long-chain bases and a-hydroxyl fatty acids. The presence of d4t,8t-sphingadienine is essential for semaphorin 3A (Sema3A)-like activity. Herein, we examined the three neuropilin 1 (Nrp1) domains (a1a2, b1b2, or c), and found that a1a2 binds to d4t,8t-kCer and possesses Sema3A-like activity. kCer binds to Nrp1 with a weak affinity of mM dissociation constant (Kd). We wondered whether bovine serum albumin could influence the ligand-receptor interaction that a1a2 has with a single high affinity binding site for kCer (Kd in nM range). In the present study we demonstrated the influence of bovine serum albumin. Thermal denaturation indicates that the a1a2 domain may include intrinsically disordered region (IDR)-like flexibility. A potential interaction site on the a1 module was explored by molecular docking, which revealed a possible Nrp1 activation mechanism, in which kCer binds to Site A close to the Sema3A-binding region of the a1a2 domain. The a1 module then accesses a2 as the IDR-like flexibility becomes ordered via kCer-induced protein rigidity of a1a2. This induces intramolecular interaction between a1 and a2 through a slight change in protein secondary structure.
Keywords: ceramide; endoglycoceramidase; konjac; neurite outgrowth; neuropilin 1; semaphorin3A; sphingadienine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

