Detection of MET Alterations Using Cell Free DNA and Circulating Tumor Cells from Cancer Patients
- PMID: 32102486
- PMCID: PMC7072825
- DOI: 10.3390/cells9020522
Detection of MET Alterations Using Cell Free DNA and Circulating Tumor Cells from Cancer Patients
Abstract
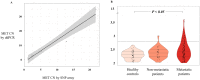
MET alterations may provide a potential biomarker to evaluate patients who will benefit from treatment with MET inhibitors. Therefore, the purpose of the present study is to investigate the utility of a liquid biopsy-based strategy to assess MET alterations in cancer patients. We analyzed MET amplification in circulating free DNA (cfDNA) from 174 patients with cancer and 49 healthy controls and demonstrated the accuracy of the analysis to detect its alteration in patients. Importantly, a significant correlation between cfDNA concentration and MET copy number (CN) in cancer patients (r = 0.57, p <10-10) was determined. Furthermore, we evaluated two approaches to detect the presence of MET on circulating tumor cells (CTCs), using the CellSearch® and Parsortix systems and monitored patients under anti-EGFR treatment (n = 30) combining both cfDNA and CTCs analyses. This follow-up provides evidence for the potential of MET CN assessment when patients develop resistance to anti-EGFR therapy and a significant association between the presence of CTCs MET+ and the Overall Survival (OS) in head and neck cancer patients (P = 0.05; HR = 6.66). In conclusion, we develop specific and noninvasive assays to monitor MET status in cfDNA/CTCs and demonstrate the utility of plasma MET CN determination as a biomarker for monitoring the appearance of resistance to anti-EGFR therapy.
Keywords: MET amplification; MET copy number; MET protein expression; circulating free DNA (cfDNA); circulating tumor cells (CTCs); targeted therapy.
Conflict of interest statement
Rafael López-López reports grants and personal fees from Roche, Merck, AstraZeneca, Bayer, Pharmamar, Leo, and personal fees and non-financial support from Bristol-Myers Squibb and Novartis, outside of the submitted work. Jorge Garcia-González reports personal fees from Novartis, Bristol-Myers Squibb, MSD Oncology, Roche/Genentech, Lilly, Boehringer Ingelheim, AstraZeneca, Pierre Fabre, Leo Pharma, and Travel, Accommodations and Expenses from Bristol-Myers Squibb, MSD Oncology, Roche/Genentech, and Lilly. Luis León-Mateos reports personal fees from Astra Zeneca, Boehringer, MSD, Sanofi, Novartis, Roche, and Bristol-Myers outside of the submitted work.
Figures








Similar articles
-
A Prospective Evaluation of Circulating Tumor Cells and Cell-Free DNA in EGFR-Mutant Non-Small Cell Lung Cancer Patients Treated with Erlotinib on a Phase II Trial.Clin Cancer Res. 2016 Dec 15;22(24):6010-6020. doi: 10.1158/1078-0432.CCR-16-0909. Epub 2016 Jun 8. Clin Cancer Res. 2016. PMID: 27281561 Clinical Trial.
-
Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy.Int J Mol Sci. 2017 May 11;18(5):1035. doi: 10.3390/ijms18051035. Int J Mol Sci. 2017. PMID: 28492516 Free PMC article.
-
Comprehensive characterization of circulating tumor cells and cell-free DNA in patients with metastatic melanoma.Mol Oncol. 2024 Nov;18(11):2770-2782. doi: 10.1002/1878-0261.13650. Epub 2024 May 24. Mol Oncol. 2024. PMID: 38790134 Free PMC article.
-
Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients.Lung Cancer. 2017 May;107:100-107. doi: 10.1016/j.lungcan.2016.04.026. Epub 2016 May 4. Lung Cancer. 2017. PMID: 27180141 Review.
-
Circulating tumour DNA: A new biomarker to monitor resistance in NSCLC patients treated with EGFR-TKIs.Biochim Biophys Acta Rev Cancer. 2020 Apr;1873(2):188363. doi: 10.1016/j.bbcan.2020.188363. Epub 2020 Apr 8. Biochim Biophys Acta Rev Cancer. 2020. PMID: 32275933 Review.
Cited by
-
The Overview of Perspectives of Clinical Application of Liquid Biopsy in Non-Small-Cell Lung Cancer.Life (Basel). 2022 Oct 19;12(10):1640. doi: 10.3390/life12101640. Life (Basel). 2022. PMID: 36295075 Free PMC article. Review.
-
Relevance of Circulating Tumor Cells as Predictive Markers for Cancer Incidence and Relapse.Pharmaceuticals (Basel). 2022 Jan 6;15(1):75. doi: 10.3390/ph15010075. Pharmaceuticals (Basel). 2022. PMID: 35056131 Free PMC article. Review.
-
Current Status and Future Perspectives of Liquid Biopsy in Small Cell Lung Cancer.Biomedicines. 2021 Jan 7;9(1):48. doi: 10.3390/biomedicines9010048. Biomedicines. 2021. PMID: 33430290 Free PMC article. Review.
-
The Development and Role of Capmatinib in the Treatment of MET-Dysregulated Non-Small Cell Lung Cancer-A Narrative Review.Cancers (Basel). 2023 Jul 10;15(14):3561. doi: 10.3390/cancers15143561. Cancers (Basel). 2023. PMID: 37509224 Free PMC article. Review.
-
The Role of MET in Resistance to EGFR Inhibition in NSCLC: A Review of Mechanisms and Treatment Implications.Cancers (Basel). 2023 May 31;15(11):2998. doi: 10.3390/cancers15112998. Cancers (Basel). 2023. PMID: 37296959 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

