Genomic Investigation Reveals Contaminated Detergent as the Source of an Extended-Spectrum-β-Lactamase-Producing Klebsiella michiganensis Outbreak in a Neonatal Unit
- PMID: 32102855
- PMCID: PMC7180233
- DOI: 10.1128/JCM.01980-19
Genomic Investigation Reveals Contaminated Detergent as the Source of an Extended-Spectrum-β-Lactamase-Producing Klebsiella michiganensis Outbreak in a Neonatal Unit
Abstract
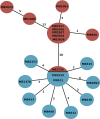
Klebsiella species are problematic pathogens in neonatal units and may cause outbreaks, for which the sources of transmission may be challenging to elucidate. We describe the use of whole-genome sequencing (WGS) to investigate environmental sources of transmission during an outbreak of extended-spectrum-β-lactamase (ESBL)-producing Klebsiella michiganensis colonizing neonates. Ceftriaxone-resistant Klebsiella spp. isolated from neonates (or their mothers) and the hospital environment were included. Short-read sequencing (Illumina) and long-read sequencing (MinION; Oxford Nanopore Technologies) were used to confirm species taxonomy, to identify antimicrobial resistance genes, and to determine phylogenetic relationships using single-nucleotide polymorphism profiling. A total of 21 organisms (10 patient-derived isolates and 11 environmental isolates) were sequenced. Standard laboratory methods identified the outbreak strain as an ESBL-producing Klebsiella oxytoca, but taxonomic assignment from WGS data suggested closer identity to Klebsiella michiganensis Strains isolated from multiple detergent-dispensing bottles were either identical or closely related by single-nucleotide polymorphism comparison. Detergent bottles contaminated by K. michiganensis had been used for washing milk expression equipment. No new cases were identified once the detergent bottles were removed. Environmental reservoirs may be an important source in outbreaks of multidrug-resistant organisms. WGS, in conjunction with traditional epidemiological investigation, can be instrumental in revealing routes of transmission and guiding infection control responses.
Keywords: Klebsiella michiganensis; Klebsiella oxytoca; extended-spectrum β-lactamase; outbreak; whole-genome sequencing.
Copyright © 2020 American Society for Microbiology.
Figures

References
-
- Stapleton PJ, Murphy M, McCallion N, Brennan M, Cunney R, Drew RJ. 2016. Outbreaks of extended spectrum beta-lactamase-producing Enterobacteriaceae in neonatal intensive care units: a systematic review. Arch Dis Child Fetal Neonatal Ed 101:F72–F78. doi:10.1136/archdischild-2015-308707. - DOI - PubMed
-
- Founou RC, Founou LL, Allam M, Ismail A, Essack SY. 2018. Genomic characterisation of Klebsiella michiganensis co-producing OXA-181 and NDM-1 carbapenemases isolated from a cancer patient in uMgungundlovu District, KwaZulu-Natal Province, South Africa. S Afr Med J 109:7–8. doi:10.7196/SAMJ.2018.v109i1.13696. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

