Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions
- PMID: 32102938
- PMCID: PMC7045384
- DOI: 10.1128/mSphere.00027-20
Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions
Abstract
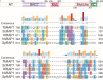
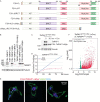
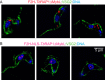
RAP1 is a telomere protein that is well conserved from protozoa to mammals. It plays important roles in chromosome end protection, telomere length control, and gene expression/silencing at both telomeric and nontelomeric loci. Interaction with different partners is an important mechanism by which RAP1 executes its different functions in yeast. The RAP1 ortholog in Trypanosoma brucei is essential for variant surface glycoprotein (VSG) monoallelic expression, an important aspect of antigenic variation, where T. brucei regularly switches its major surface antigen, VSG, to evade the host immune response. Like other RAP1 orthologs, T. brucei RAP1 (TbRAP1) has conserved functional domains, including
Keywords: RAP1; Trypanosoma brucei; protein-protein interaction; telomere.
Copyright © 2020 Afrin et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
