DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis
- PMID: 32103174
- PMCID: PMC10919329
- DOI: 10.1038/s41586-020-2041-2
DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis
Abstract
The DNA-dependent protein kinase (DNA-PK), which comprises the KU heterodimer and a catalytic subunit (DNA-PKcs), is a classical non-homologous end-joining (cNHEJ) factor1. KU binds to DNA ends, initiates cNHEJ, and recruits and activates DNA-PKcs. KU also binds to RNA, but the relevance of this interaction in mammals is unclear. Here we use mouse models to show that DNA-PK has an unexpected role in the biogenesis of ribosomal RNA (rRNA) and in haematopoiesis. The expression of kinase-dead DNA-PKcs abrogates cNHEJ2. However, most mice that both expressed kinase-dead DNA-PKcs and lacked the tumour suppressor TP53 developed myeloid disease, whereas all other previously characterized mice deficient in both cNHEJ and TP53 expression succumbed to pro-B cell lymphoma3. DNA-PK autophosphorylates DNA-PKcs, which is its best characterized substrate. Blocking the phosphorylation of DNA-PKcs at the T2609 cluster, but not the S2056 cluster, led to KU-dependent defects in 18S rRNA processing, compromised global protein synthesis in haematopoietic cells and caused bone marrow failure in mice. KU drives the assembly of DNA-PKcs on a wide range of cellular RNAs, including the U3 small nucleolar RNA, which is essential for processing of 18S rRNA4. U3 activates purified DNA-PK and triggers phosphorylation of DNA-PKcs at T2609. DNA-PK, but not other cNHEJ factors, resides in nucleoli in an rRNA-dependent manner and is co-purified with the small subunit processome. Together our data show that DNA-PK has RNA-dependent, cNHEJ-independent functions during ribosome biogenesis that require the kinase activity of DNA-PKcs and its phosphorylation at the T2609 cluster.
Conflict of interest statement
Figures





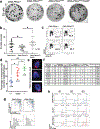




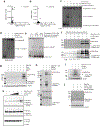
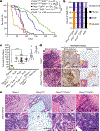
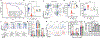


Comment in
-
DNA-repair enzyme turns to translation.Nature. 2020 Mar;579(7798):198-199. doi: 10.1038/d41586-020-00424-7. Nature. 2020. PMID: 32152600 No abstract available.
References
-
- Zhu C et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109, 811–821 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

