Pathway paradigms revealed from the genetics of inflammatory bowel disease
- PMID: 32103191
- PMCID: PMC7871366
- DOI: 10.1038/s41586-020-2025-2
Pathway paradigms revealed from the genetics of inflammatory bowel disease
Abstract
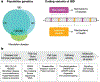
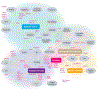
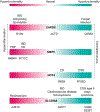
Inflammatory bowel disease (IBD) is a complex genetic disease that is instigated and amplified by the confluence of multiple genetic and environmental variables that perturb the immune-microbiome axis. The challenge of dissecting pathological mechanisms underlying IBD has led to the development of transformative approaches in human genetics and functional genomics. Here we describe IBD as a model disease in the context of leveraging human genetics to dissect interactions in cellular and molecular pathways that regulate homeostasis of the mucosal immune system. Finally, we synthesize emerging insights from multiple experimental approaches into pathway paradigms and discuss future prospects for disease-subtype classification and therapeutic intervention.
Conflict of interest statement
Competing interests
R.J.X. is a consultant to Novartis and Nestle.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

