Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification
- PMID: 32103255
- PMCID: PMC7144905
- DOI: 10.1093/nar/gkaa099
Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification
Abstract
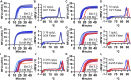
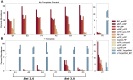
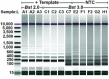
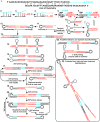
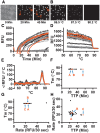
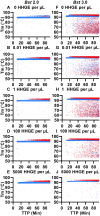
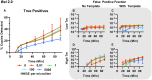
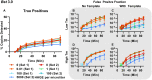
Isothermal amplification assays, such as loop-mediated isothermal amplification (LAMP), show great utility for the development of rapid diagnostics for infectious diseases because they have high sensitivity, pathogen-specificity and potential for implementation at the point of care. However, elimination of non-specific amplification remains a key challenge for the optimization of LAMP assays. Here, using chlamydia DNA as a clinically relevant target and high-throughput sequencing as an analytical tool, we investigate a potential mechanism of non-specific amplification. We then develop a real-time digital LAMP (dLAMP) with high-resolution melting temperature (HRM) analysis and use this single-molecule approach to analyze approximately 1.2 million amplification events. We show that single-molecule HRM provides insight into specific and non-specific amplification in LAMP that are difficult to deduce from bulk measurements. We use real-time dLAMP with HRM to evaluate differences between polymerase enzymes, the impact of assay parameters (e.g. time, rate or florescence intensity), and the effect background human DNA. By differentiating true and false positives, HRM enables determination of the optimal assay and analysis parameters that leads to the lowest limit of detection (LOD) in a digital isothermal amplification assay.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Kong J.E., Wei Q., Tseng D., Zhang J., Pan E., Lewinski M., Garner O.B., Ozcan A., Di Carlo D.. Highly stable and sensitive nucleic acid amplification and cell-phone-based readout. ACS Nano. 2017; 11:2934–2943. - PubMed
-
- Somboonna N., Choopara I., Arunrut N., Sukhonpan K., Sayasathid J., Dean D., Kiatpathomchai W.. Rapid and sensitive detection of Chlamydia trachomatis sexually transmitted infections in resource-constrained settings in Thailand at the point-of-care. PLoS Negl. Trop. Dis. 2018; 12:e0006900. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

