Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli
- PMID: 32106218
- PMCID: PMC8142898
- DOI: 10.1038/s41586-020-2080-8
Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli
Abstract
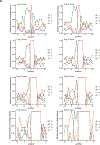
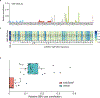
Various species of the intestinal microbiota have been associated with the development of colorectal cancer1,2, but it has not been demonstrated that bacteria have a direct role in the occurrence of oncogenic mutations. Escherichia coli can carry the pathogenicity island pks, which encodes a set of enzymes that synthesize colibactin3. This compound is believed to alkylate DNA on adenine residues4,5 and induces double-strand breaks in cultured cells3. Here we expose human intestinal organoids to genotoxic pks+ E. coli by repeated luminal injection over five months. Whole-genome sequencing of clonal organoids before and after this exposure revealed a distinct mutational signature that was absent from organoids injected with isogenic pks-mutant bacteria. The same mutational signature was detected in a subset of 5,876 human cancer genomes from two independent cohorts, predominantly in colorectal cancer. Our study describes a distinct mutational signature in colorectal cancer and implies that the underlying mutational process results directly from past exposure to bacteria carrying the colibactin-producing pks pathogenicity island.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures




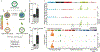
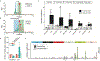
Comment in
-
Colibactin Causes Colorectal Cancer-Associated Mutational Signature.Cancer Discov. 2020 May;10(5):635. doi: 10.1158/2159-8290.CD-RW2020-037. Epub 2020 Mar 13. Cancer Discov. 2020. PMID: 32169880
-
Leaving your mark.Nat Rev Cancer. 2020 May;20(5):260. doi: 10.1038/s41568-020-0254-1. Nat Rev Cancer. 2020. PMID: 32203301 No abstract available.
-
A mutational signature that can be made by a bacterium arises in human colon cancer.Nature. 2020 Apr;580(7802):194-195. doi: 10.1038/d41586-020-00818-7. Nature. 2020. PMID: 32203380 No abstract available.
-
Microbiota and colorectal cancer: colibactin makes its mark.Nat Rev Gastroenterol Hepatol. 2020 Jun;17(6):317-318. doi: 10.1038/s41575-020-0303-y. Nat Rev Gastroenterol Hepatol. 2020. PMID: 32317778 No abstract available.
-
Genetic Evidence That a Gut Commensal Bacterium Can Cause Colorectal Cancer.Gastroenterology. 2021 Mar;160(4):1424-1426. doi: 10.1053/j.gastro.2020.12.014. Epub 2020 Dec 11. Gastroenterology. 2021. PMID: 33310089 No abstract available.
References
-
- Gagnaire A, Nadel B, Raoult D, Neefjes J & Gorvel J-P Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat. Rev. Microbiol. 15, 109–128 (2017). - PubMed
-
- Nougayrède J-P et al. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 313, 848–851 (2006). - PubMed
REFERENCES FROM METHODS SECTION
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

