Chromothripsis and DNA Repair Disorders
- PMID: 32106411
- PMCID: PMC7141117
- DOI: 10.3390/jcm9030613
Chromothripsis and DNA Repair Disorders
Abstract
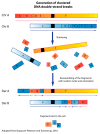
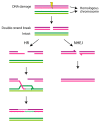
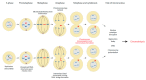
Chromothripsis is a mutational mechanism leading to complex and relatively clustered chromosomal rearrangements, resulting in diverse phenotypic outcomes depending on the involved genomic landscapes. It may occur both in the germ and the somatic cells, resulting in congenital and developmental disorders and cancer, respectively. Asymptomatic individuals may be carriers of chromotriptic rearrangements and experience recurrent reproductive failures when two or more chromosomes are involved. Several mechanisms are postulated to underlie chromothripsis. The most attractive hypothesis involves chromosome pulverization in micronuclei, followed by the incorrect reassembly of fragments through DNA repair to explain the clustered nature of the observed complex rearrangements. Moreover, exogenous or endogenous DNA damage induction and dicentric bridge formation may be involved. Chromosome instability is commonly observed in the cells of patients with DNA repair disorders, such as ataxia telangiectasia, Nijmegen breakage syndrome, and Bloom syndrome. In addition, germline variations of TP53 have been associated with chromothripsis in sonic hedgehog medulloblastoma and acute myeloid leukemia. In the present review, we focus on the underlying mechanisms of chromothripsis and the involvement of defective DNA repair genes, resulting in chromosome instability and chromothripsis-like rearrangements.
Keywords: DNA double-strand breaks (DSBs); DNA repair; DNA repair disorders; TP53; ataxia telangiectasia and Rad3-related (ATR); ataxia telangiectasia mutated (ATM); chromosome pulverization; chromothripsis; micronuclei; structural variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

